Major advancements in both treatment and diagnostic technologies for HIV infection have occurred during the last 20 years. Life expectancy for a person infected with HIV, if he or she is diagnosed early and treated promptly, is approaching that of uninfected individuals.1 In many respects HIV infection can now be considered a “chronic illness.” Attitudes towards HIV testing have also changed. Increasingly, HIV testing has become accepted as part of routine medical care.
Despite these encouraging developments, a substantial proportion of HIV infections remain undiagnosed in the United States. It has been estimated that 20% of the 1.2 million HIV-infected individuals in the U.S. are unaware of their HIV positive status.2 Undiagnosed persons are more likely to continue to participate in high-risk behavior, cannot benefit from treatment, and disproportionately account for new HIV infections. Today, the emphasis is on improving access to testing, early diagnosis, and streamlining referral into care.
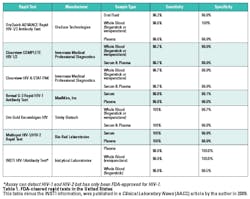
Many advances in HIV diagnostic technologies have made these goals more attainable. A key development was the availability of rapid tests that can be used in a variety of non-traditional settings. There are seven FDA-approved rapid tests in the U.S. (Table 1). Rapid tests are performed on a variety of different sample types. They are significantly more expensive than traditional enzyme immunoassays but provide results in less than 30 minutes. The latest addition, INSTI (bioLytical Laboratories), produces results in only 60 seconds. Assays that use unprocessed sample types such as whole blood and oral fluid are CLIA waived; the remainder are categorized as moderately complex. The majority, but not all, can detect both HIV-1 and HIV-2 antibodies. All rapid tests have sensitivity and specificity values greater than 99.0% for detection of antibodies to HIV-1.3,4 Of note, only Multispot has the ability to not only detect but also discriminate between HIV-1 and HIV-2 infection. This year also marks the approval of the first over-the-counter, rapid HIV test for home use, OraQuick (OraSure Technologies).
Another milestone in HIV diagnostics was the approval of HIV Ag/Ab combination or fourth generation assays. There are two assays in the U.S. market that have this capability. The primary advantage of the Antigen/Antibody (Ag/Ab) combination assays is that they can detect HIV infection on average five to seven days earlier than third-generation antigen sandwich assays that only detect antibodies. Reducing the seroconversion window has always been an important goal in HIV diagnostics, as it has long been recognized that individuals with acute HIV infection have peak viral loads and are highly infectious.5
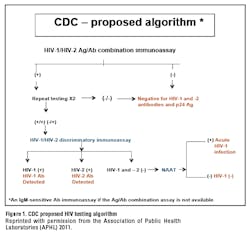
The recent availability of diagnostic assays that can detect HIV infection earlier has prompted a new HIV diagnostic algorithm that has been proposed by the CDC and officially published in the CLSI (Clinical and Laboratory Standards Institute) document M-53A: Criteria for Laboratory Testing and Diagnosis of Human Immunodeficiency Virus Infection: Approved Guideline (Figure 1). Although the CDC algorithm is only one of several algorithms presented in the CLSI document, the CDC version has generated the most interest because it represents a fundamental change to current practice. The two-step, immunoassay (IA)/rapid test screen followed by Western blot confirmation approach that has been the gold standard for HIV diagnosis for more than two decades is no longer the preferred strategy. The proposed algorithm promises to improve, first, the detection of acute HIV infection, and, second, differentiation between HIV-1 and HIV-2 infection, all with faster turnaround time. This article reviews issues with the current algorithm, highlights some of the more recent data in support of the alternative algorithm, and addresses practical aspects of adopting the new algorithm. From the laboratory perspective, the proposed algorithm provides new opportunities but also a few challenges, including limited assay options at key points in the algorithm.
THE NEW ALGORITHM
The first step: Ag/Ab combination immunoassay
The first step in the proposed algorithm is the recommendation of an Ag/Ab combination HIV immunoassay, also referred to as fourth-generation assay, as the initial screening test performed in the laboratory. HIV Ag/Ab combination assays simultaneously detect HIV-1 (groups M, N, O) and HIV-2 antibodies, as well as p24 antigen. The major advantage of the fourth generation assays as compared to third generation Ab only assays is the improved ability to identify acute HIV infection (AHI) prior to seroconversion, due to detection of p24 antigen. However, nucleic acid amplification test (NAAT) is the most sensitive methodology for detecting acute HIV infection.
Currently there are only two FDA-approved HIV Ag/Ab combination assays in the U.S. The ARCHITECT HIV Ag/Ab Combo Assay (Abbott Laboratories) is offered on a large, fully automated instrument. The GS HIV Combo Ag/Ab EIA (Bio-Rad Laboratories) is a microplate-based assay that can be performed manually or using a benchtop platform. While increased sensitivity for acute HIV infection is important, it’s key that these assays also retain sensitivity for HIV-1 (groups M and O) and HIV-2 established infections, as well as specificity in low-risk populations. Chavez’s group evaluated almost 11,000 samples and demonstrated that the sensitivity and specificity were 99.94% and 99.50% respectively for the ARCHITECT Ag/Ab Combo assay. This assay detected 83% of acute HIV specimens.6
Bentsen and colleagues determined that the GS HIV Combo Ag/Ab EIA sensitivity was 100% with known antibody positive HIV-1 (groups M and O) and HIV-2 samples. The assay also detected 95% and 86% of acute HIV samples in the two populations studied. In addition, the p24 antigen was detected in 100% of cases using supernatants from groups M, (B and non-B subtypes included) N, and O. Analytical sensitivity for p24 antigen utilizing a WHO/NIBSC HIV international standard was
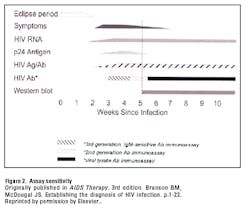
Although there is a clearly stated preference for Ag/Ab combination assays as the initial screening assay, the CLSI guidelines do make an allowance for the use of third-generation antibody assays. Third-generation assays typically have an antigen bridging format and detect both IgG and IgM antibodies. HIV infection can be diagnosed at approximately 22 days post infection using these assays (Figure 2). In contrast to Ag/Ab combination assays, third- generation assays are readily available from a number of vendors on large, fully automated immunoassay analyzers and have been implemented by most laboratories.
The second step: HIV-1/HIV-2 discriminatory assay
The proposed algorithm eliminates the Western blot and recommends the use of an assay that can discriminate between HIV-1 and HIV-2 infection as the second supplemental test.
The Multispot. In practice, the only FDA-approved assay that can discriminate between HIV-1 and HIV-2 antibodies and meets the criteria for the second test in the algorithm is the Multispot (Bio-Rad). This unique attribute is the primary reason for replacing the Western blot with the Multispot. The Multispot is a rapid antibody test that is categorized as moderately complex and can be performed in a laboratory on plasma and serum samples. Studies have demonstrated that the Multispot is less sensitive than third-generation and fourth-generation screening assays, as expected, but slightly more sensitive than the Western blot (WB).8,9 The sensitivity and specificity of Multispot are 100.0% and 99.9%, respectively, for HIV-1 antibodies. Because the Multispot is a second generation rapid test (detects IgG antibodies), it detects infection in individuals who have been infected longer than three months and have seroconverted, who comprise the majority of HIV diagnoses. Specimens that are negative at this point proceed to a nucleic acid amplification test (NAAT) to rule out acute HIV infection. An added benefit to the Multispot is that its rapid test format reduces turnaround time and expedites HIV diagnosis. One complication is that the Multispot was not specifically approved as a supplemental test for HIV-1 diagnosis.
Eliminating the Western blot. Indirect immunofluroescence assay (IFA) is also FDA-approved for confirming HIV-1 infection, but because the Western blot is used by more than 98% of laboratories in the U.S., it has become synonymous with HIV confirmation. Although progressive advances have been made in HIV screening assays, the Western blot confirmatory assay has not been modified since its original development in the late 1980s. The result is that screening assays have outpaced the Western blot in terms of sensitivity, because the Western blot can still only detect IgG antibodies (Figure 2). For example, the lower sensitivity of Western blot during the seroconversion period can yield indeterminate or negative confirmatory results in persons who were correctly classified as HIV-infected by third-generation screening assays. This problem is bound to be exacerbated by Ag/Ab combination assays that further increase the sensitivity gap between screening assays and confirmation. These findings were recently confirmed by a comprehensive study that evaluated six rapid tests, one fourth-generation and three third-generation immunoassays, Western blot, and one NAAT for detection of HIV infection.8 In light of the increased sensitivity of the fourth-generation assays, using the Western blot alone for confirmation is clearly inadequate.
Another limitation to using the Western blot as a confirmatory assay is that the Western blot was designed for confirmation of HIV-1 infection. Here again, unlike the Western blot, screening assay technologies have evolved rapidly to incorporate antigens for detection of infection with rare HIV strains, including HIV-1 group O and HIV-2. Both fourth-generation and third-generation assays, as well as many rapid tests, have the ability to detect HIV-1 and HIV-2 antibodies. HIV-2 infection is rare and, according to the CDC, only 166 cases met the criteria for HIV-2 infection between the years 1988 and 2010 in the U.S. Eighty-one percent (81%) of these patients were born in West Africa.10 HIV-2 specific testing has been recommended for patients with epidemiologic links to West Africa, individuals with opportunistic infections suggestive of HIV infection who are negative for HIV-1 infection, or patients with a distinctive indeterminate Western blot pattern that consists of presence of gag (p55, p24) and pol (p66, p51, p31) but absence of env bands.
Missing and/or misclassifying HIV-2 infection by using the Western blot as the confirmatory assay has been a concern. Recent evidence in support of this concern was published by Nasrullah and colleagues, who investigated the Western blot patterns and interpretations for 1761 known HIV-1 and 114 HIV-2 samples. The pattern observed for HIV-2 infection was consistent with previously published reports. Using the CDC criteria for interpretation of Western blots, of 114 known HIV-2 samples, 1 (0.9) was categorized as negative, 60 (52.6%) as indeterminate and, importantly, 53 (46.5%) were categorized as positive for HIV-1 infection.11 The consequences of missing HIV-2 infection (negative and indeterminate results) are clear, but misclassifying HIV-2 as HIV-1 also has important clinical implications. Many first-line drugs used to treat HIV-1 infection are ineffective for treating HIV-2, and furthermore NAAT assays are optimized for detecting HIV-1 infections and cannot be used to effectively detect or monitor HIV-2 infections. Available assays for detecting HIV-2 infection include HIV-2 specific EIA, HIV-2 immunoblot, and HIV-2 DNA and RNA specific NAAT tests. Many of these are laboratory developed tests. There is currently no FDA-approved assay for confirmation of HIV-2 infection.
Lastly, although the Western blot is prized for its specificity when a result is positive, indeterminate results are encountered. An indeterminate Western blot can be the result of many causes. These include HIV-2 infection, acute HIV infection with incomplete seroconversion, late stage disease (AIDS) with immunosuppression, HIV vaccinated individuals, cross-reactivity with other viral or cellular components, and other infections.12 An indeterminate Western blot result invariably necessitates follow-up to distinguish among these possibilities, although it rarely indicates HIV infection.
Practical considerations that limit the use of Western blot include expense, the frequent need to refer the sample to a reference laboratory for testing, and extended turnaround time.
The third step: Nucleic Acid Amplification Tests (NAATs)
For the first time, HIV-1 RNA assays are formally included in the diagnostic algorithm to resolve any discrepant results between the first and second steps in the algorithm. NAATs are the most sensitive assays for detection of HIV infection (Figure 2). NAATs require extensive expertise, relatively expensive reagents and platforms, and a reasonable test volume. HIV RNA molecular assays are either qualitative or quantitative. Quantitative assays are referred to as viral load (VL) assays and are primarily reserved for prognosis and monitoring treatment response. Much progress has been made in viral load assays, including the development of real-time PCR assays. These important advances have led to VL assays with broader dynamic range and coverage for rare HIV genotypes, and analytical sensitivity ranges as low as 20 to 50 copies/mL, depending on the assay. Quantitative assays are available on a variety of high-throughput platforms and are widely utilized.
Despite the significant strides made in VL performance characteristics, the limitation is that these assays are not FDA-approved for diagnostic use. Therefore, although potentially clinically useful as both supplemental diagnostic tests and patient management tools, utilizing these as diagnostic assays would constitute “off-label” use. Offering these as supplemental tests would require either that vendors seek additional FDA approval for a diagnostic claim or laboratories perform validation studies that would meet regulatory requirements. On the other hand, the APTIMA HIV-1 RNA Qualitative Assay (Gen-Probe) is not only approved for confirmation of repeatedly reactive immunoassay results but also for diagnosis of acute HIV infection. APTIMA, however, is a semi-automated assay and few laboratories offer this type of testing.
Importantly, there are true cases of HIV infection that are negative by nucleic acid amplification tests. Approximately 0.5% of HIV infected individuals, known as elite suppressors/controllers, can have undetectable viral loads in the absence of treatment. Other possibilities include uninfected babies born to HIV-positive mothers and HIV-2 infection. Current NAATs are not designed to detect HIV-2 infection, although some laboratories have developed in-house HIV-2 DNA and RNA assays. Molecular assays for detection of HIV-2 infection are rare in the U.S. Outside the U.S., where these assays are more common, much variation exists among results from different laboratories.13
Summary
In conclusion, important advances have been made for diagnosis of HIV infection. It’s clear that early diagnosis and prompt treatment improve outcome and reduce disease transmission. New tools are now available that substantially enhance detection of HIV infection. The rapid advances in technology, however, necessitate new strategies for diagnosis. The algorithm proposed by the CDC incorporates new and old assays in a test sequence that maximizes effectiveness. The proposed algorithm is predicated on data from several studies, many of which were published in a special issue of Clinical Virology, December 2011. Individual assays, as well as algorithms with various test components, were evaluated for specificity and sensitivity for both established and primary HIV infections. Masciotra and colleagues determined that there were no significant differences in sensitivity and specificity between the conventional two-test and the proposed three-test algorithms.
Overall the data support the new algorithm, as it increases detection of acute HIV infections and improves discrimination between HIV-1 and HIV-2 infections, without significant reduction in specificity or sensitivity for established HIV-1 infections. The new HIV diagnostic algorithm is also designed to use already FDA-approved HIV assays available in the U.S., although the number of options is very limited at certain key points in the algorithm. Testing volumes, cost of new or automated instrumentation, and the population that is served by a laboratory are all variables that will influence the ability of an institution to implement a particular assay and algorithm. For example, if a laboratory serves a high incidence/high prevalence population, then emphasis is on detection of acute HIV infection, and a fourth-generation assay will be necessary. If a laboratory has very low testing volumes, then investing in and/or maintaining a highly automated platform may not be feasible, and sending testing to a reference laboratory would be preferable. In reality, adoption of the new algorithm will take time as many laboratories will wrestle with the expense and logistics of purchasing new instrumentation and validating the recommended assays. Formal recommendations from CDC providing further guidance to laboratories are eagerly anticipated, and it is hoped that they will be released before the end of 2012. As laboratorians we want to adopt new technologies, especially when they benefit the patient and society, but we must also be informed of the regulatory and practical aspects of offering new assays.
References
- van Sighem AI, Gras LA, Reiss P, Brinkman K, de Wolf F, ANOC study. Life expectancy of recently diagnosed asymptomatic HIV-infected patients approaches that of uninfected individuals. AIDS. 2010;24(10):1527-1535.
- Centers for Disease Control and Prevention. HIV surveillance—United States 1981-2008. MMWR. 2011;60(21):689-693.
- bioLytical Laboratories Inc, INSTI (package insert). Richmond, BC, Canada. 2010.
- U.S. Department of Health and Human Services. General and laboratory considerations: rapid HIV tests curently available in the United States. 2007.
http://www.cdc.gov/hiv/topics/testing/resources/factsheets/rt-lab.htm. Accessed September 24, 2012. - Brenner BG, Roger M, Routy JP, et al. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis. 2007;195(7):951-959.
- Chavez P, Wesolowski L, Patel P, Delaney K, Owen SM. Evaluation of the performance of the Abbott ARCHITECT HIV Ag/Ab Combo Assay. J Clin Virol. 2011;52(1):51-55.
- Bentsen C, McLaughlin L, Mitchell E, et al. Performance evaluation of the Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA, a 4th generation HIV assay for the simultaneous detection of HIV p24 antigen and antibodies to HIV-1 (groups M and O) and HIV-2 in human serum or plasma. J Clin Virol. 2011;52(1):57-61.
- Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(1):17-22.
- Torian LV, Forgione LA, Punsalang AE, Pirillo RE, Oleszko WR. Comparison of Multispot EIA with Western blot for confirmatory serodiagnosis of HIV. J Clin Virol. 2011;52(1):41-44.
- Centers for Disease Control and Prevention. HIV-2 Infection Surveilance – United States, 1987-2009. MMWR. 2011;60(29):985-988.
- Nasrullah M, Ethridge SF, Delaney KP, et al. Comparison of alternative interpretive criteria for the HIV-1 Western blot and results of the Multispot HIV-1/HIV-2 Rapid Test for classifying HIV-1 and HIV-2 infections. J Clin Virol. 2011;52(1):S23-27.
- Mahajan VS, Pace CA, Jarolim P. Interpretation of HIV serologic testing results. Clin Chem. 2010;56(10):1523-1526.
- Damond F, Benard A, Ruelle J, et al. Quality control assessment of human immunodeficiency virus type 2 (HIV-2) viral load quantification assays: results from an international collaboration on HIV-2 infection in 2006. J Clin Microbiol. 2008;46(6):2088-2091.