The anarchy of antibiotic resistance: mechanisms of bacterial resistance
To earn CEUs, visit mlo-online.com and see this month’s test by clicking the CE menu item.
Sponsored
by Abbott
Diagnostics
LEARNING OBJECTIVES
Upon completion of this article, the reader will be able to:
- discuss Gram-positive versus Gram-negative resistance;
- describe the features of NDM-1 and its transmission;
- discuss the identification of ESBL, KPC, and MBL enzymes;
- explain mechanisms of antibiotic resistance; give examples; and
- discuss the global concerns of epidemiology regarding ESBLs and MBLs.
From Staphylococcus aureus resistance to penicillin in the 1940s and methicillin resistance in the 1960s, the focus of antibiotic resistance has been on Gram-positive bacteria. Glaring reports of methicillin-resistant S aureus (MRSA) infection aroused public awareness and brought research priorities to the forefront in the 1970s and 1980s. Yet another exigency arrived in the 1990s with the increasing appearance of the extended-spectrum b-lactamases (ESBLs), enzymes carried by Gram-negative bacteria belonging to the Enterobacteriacae family. These multidrug-resistant (MDR) bacteria have created devastating disease, both hospital-acquired (HA) and community-acquired (CA), and are of greatest concern to infectious-disease specialists and public-health officials, worldwide.
Gram-negative bacteria have the potential not only to develop chromosomal resistance but also to quickly spread resistance via genetic determinants carried on plasmids or transposons (i.e., pieces of DNA that jump from one strain or species to another). Rapid spread through human travel and migration, combined with the looming dilemma of “no new drugs,” these resistant elements have created a global treatment crisis — greater than from Gram-positive bacteria.
Reported first in India in the 1990s, the ESBL gene blaCTX-M-15 jumped from chromosome to plasmid through a process that expedited its worldwide spread. The CTX-M15 enzyme, which continues to be the dominant ESBL, is responsible for acquired resistance to third-generation cephalosporins in the Enterobacteriacae. In India, the presence of 70% to 90% of resistant ESBLs made treatment with carbapenem antibiotics mandatory. Most frightening, the once successful ESBL therapy has resulted in selection for resistance to the carbapenems, leaving few alternatives in the antibiotic armamentarium.1
A large study in 2010 identified more than 150 isolates of a new metallo-b-lactamase (MBL), named NDM-1 from India, the U.K., and Pakistan. The characteristic gene, blaNDM-1, is carried on plasmids and easily transferred among many bacteria, in a manner similar to the spread of the CTX-M15. This typical and flexible type of transmission creates tremendous potential for the rapid spread of resistant genes throughout the environment. Carbapenem-resistant Klebsiella pneumoniae and plasmids that encode Verona integron-encoded MBL(VIM) in K pneumoniae, already found in the U.S. Greece, Israel, et al, are responsible for escalating the present healthcare crisis. 1
Arrival of NDM-1
In 2007, a zinc-dependent MBL gene, blaNDM-1, was isolated in New Delhi, India, from the urine culture of a 59-year-old patient who had type II diabetes and a history of strokes. Although his residence was in Sweden, he often traveled to India, his home of origin. During a December 2007 trip, he was hospitalized in Punjab with a gluteal abscess. Later admitted to a New Delhi hospital (for a second surgery), he developed a decubitus ulcer (deep wound). In January 2008, he was referred to Orebro, Sweden, where he was treated parenterally with amoxacillin-clavulanic acid, metronidazole, amikacin, and gatifloxacin. At that time, a carbapenem-resistant K pneumoniae, characterized as an MBL unlike any known MBL, was isolated from a urine culture.
The new “subgroup” was designated NDM-1 or K pneumoniae 05-506, blaNDM-1, which was carried on a large plasmid (180 kb) and easily transferred to other species. The patient had not experienced symptoms of urinary-tract infection, or UTI, but the isolate exhibited resistance to all antibiotics on the Gram-negative formulary — except the fluoroquinolones and colistin. More important, the isolate was carbapenem resistant and positive by the MBL E-test (AB bioM’erieux), determined by clinical laboratory testing. (NDM-1 binds tightly to most cephalosporins and penicillins but less to the carbapenems). An ESBL-producing Escherichia coli and Acinetobacter sp., carbapenem susceptible, were isolated from his wounds, and the same E coli was recovered in a culture of otitis fluid.
He was discharged to a nursing home in March 2008, where a new urine culture grew an ESBL-producing K pneumoniae. A fecal specimen grew an NDM-1 producing E coli (on a 140-kb plasmid), suggesting an in vivo conjugation transfer from the 180-kb K pneumoniae 05-506 had taken place, but the 05-506 strain was not recovered from any subsequent cultures.2
NDM-1 is described as a “transmissible genetic element” rather than a single bacterium because of its potential to encode multiple resistance genes. Though its origin was in New Delhi, many isolates have been reported throughout India, Pakistan, Bangladesh, the U.K., the U.S., Israel, and Turkey, some susceptible only to colistin. Other determinants found in that first isolate included the broad spectrum b-lactamase, CMY-4, and genes inactivating erythromycin, rifampin, ciprofloxacin, and chloramphenicol. The same element also encoded an efflux pump, a mechanism adding further resistance along with “growth promoters” that would ensure transcription of the genes carried by the element. 2
Because of the outbreaks of carbapenem-resistant ESBL and K pneumoniae, the rapid spread and increased resistance of NDM-1 raised a critical question regarding the potential for controlling CA UTI, henceforth. Without new antibiotics and upgraded public-health measures, the fear of another “superbug” epidemic became decidedly real.3
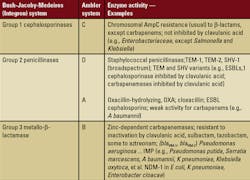
Detection and diagnosis: MBLS and KPC
Although identification of Enterobacteriaceae isolates is reliable by automated or manual (conventional) methods, antibiotic-susceptibility testing in certain resistant Gram-negative bacteria requires special processing. Presence of an ESBL or AMP-C producing isolate (i.e., resistance to first-, second-, third-, or even fourth-generation cephalosporins, plus aztreonam, ertapenem, and imipenem) may lead to misinterpreted results when tested with automated systems. Recommended instead are double-disk and combination-disk methods of detection, performed with ATCC quality-control organisms.4 [ATCC is a private, non-profit biological resource center and research organization in Manassas, VA. ATCC’s microorganism collection includes more than 18,000 strains of bacteria. Go to About ATCC for more information.]
Adapted from Rice LB, Bonomo RA. Mechanisms of Resistance to Antibacterial Agents. In:
Murray PR, Jorgensen JH,
Pfaller MA, et al, eds. Manual of Clinical Microbiology, 9th ed. Vol. 1. Washington DC: American Society for Microbiology Press; 2007:1114-1130. 1 ESBLs — extended-spectrum–lactamases.
Distinguishing the carbapenemase enzymes (Class A) produced by K pneumoniae from (Class B) MBL enzymes can be problematic. Thus, preliminary phenotypic screening by double-disk synergy and confirmation by PCR [polymerase chain reaction] are recommended (by the Centers for Disease Control and Prevention [CDC]) for detection. The double-disk method is performed with meropenem or imipenem disks (disk diffusion on Mueller-Hinton agar) alone and together with phenylboronic acid (PBA), EDTA, or PBA and EDTA together. Zone sizes augmented by ^35 mm determine a positive result for the combined-disc test. MBL bacteria are inhibited by EDTA. The test requires stringent preparation with appropriate quality controls, followed by knowledgeable interpretation of the results.4 Isolates of NDM-1 may be distinguished from K pneumoniae by PCR and collected for surveillance and epidemiologic purposes at CDC. 4
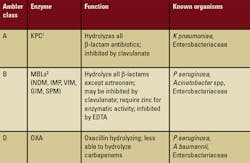
(metallo–Lactamases — MBLs)
Adapted from Rice LB, Bonomo RA. Mechanisms of Resistance to Antibacterial Agents. In: Murray PR, Jorgensen JH, Pfaller MA, et al, eds. Manual of Clinical Microbiology, 9th ed. Vol. 1 Washington DC: American Society for Microbiology Press; 2007:1114-1130.
1KPC—Klebsiella pneumoniae carbapenem-resistant
2MBL—Metallo- -lactamases (MBLs) (e.g., NDM-1, IPM, VIM, and so forth)
In the case presented here, identification of KPC 05-506 was accomplished with the Phoenix and BD automated systems. Susceptibility testing was performed with E-test strips (AB bioM’erieux). The MBL test (double-disc synergy with imipenem-EDTA) and the modified Hodge test (clover-leaf formation) were used to screen for carbapenemase production. The E-test determined susceptibility MICs [minimal inhibitory concentration] added to a spectrophotometry analysis to confirm carbapenemase activity.2
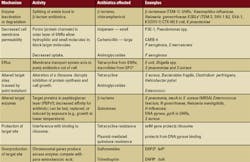
Adapted from Opal SM, Medeiros AA. Molecular mechanisms of antibiotic resistance in bacteria. In: Mandell GL, Bennett JE, Dolin R, eds. Principles and Practice of Infectious Diseases, 6th ed. Philadelphia, PA; Elsievier Churchill, Livingstone Inc. 2005;(1):253-270.
1GNRs — Gram-negative rods
2ESBLs — extended-spectrum–lactamases
3GPC — Gram-positive cocci
4PBP — penicillin-binding protein
5DHPS — dihydropteroate synthesis
6DHFR — dihydrofolate reductase
Isolates were screened by PCR for presence of the mobile MBL genes (blaVIM, blaIMP, blaSPM, blaGIM-1, blaSIM-1, blaAIM-1) and beta-lactamase genes (blaCTX, blaCMY, and so forth).Further tests included DNA cloning with restriction endonucleases, PCR amplification and sequencing, pulse field gel electrophoresis (PFGE), Southern-blot analysis, and multilocus sequence typing, or MLST. When the analysis was complete, two types of b-lactamase resistance were apparent. One fragment carried the AmpC enzyme, determined by resistance to EDTA and sensitivity to the inhibitor cloxacillin. The second fragment carried the MBL, which was resistant to the inhibitor but sensitive to EDTA in the double-disk synergy test.2,5
Development of antibiotic resistance
Intrinsic (inherent) antibiotic resistance is a known phenomenon in the diverse world of bacteria. Exposure to antibiotic agents, however, creates extreme selective pressure, forcing organisms to exert a complex array of genetic manipulation to cope and evolve in an unfavorable environment. This pressure is responsible for what we know as acquired resistance. Intrinsic resistance to vancomycin in E coli and K pneumoniae negates its use as treatment for infection, unlike the acquired low-level resistance of Streptococcus pneumoniae, which can be overridden by increasing the dosage of antibiotic (e.g., pediatric ear infection).6
Acquired antibiotic resistance is a result of biochemical encoding in the affected bacterial genes. To this end, bacteria have devised three major mechanisms of resistance: mutation of cellular genes, acquisition of resistance genes, and mutation of acquired genes. Mutation of cellular genes, or point mutation, occurs in a nucleotide base pair to alter the target site of an antibiotic, thus affecting growth and activity in the cell. The early-discovered b-lactamase genes that encode enzymes TEM-1 and SHV-1 laid the groundwork for later-recognized mutant genes encoding the ESBLs of E coli and K pneumoniae, et al. (e.g., TEM-3, SHV-2, TEM-30). An example of this type (defined as a single-point mutation) also occurs when RNA polymerase is targeted by rifampin.6
Acquisition of resistance genes by rearrangement of large segments of DNA creates a potential for independent movement from the rest of the genome. Sequences of DNA are rearranged by inversion, duplication, insertion, deletion, or transposition from one area of a bacterial chromosome or extra-chromosomal plasmid to another. The NDM-1 case presented here demonstrates a rearrangement of this type.
Other mechanisms of survival: DNA may be taken up by chromosomes via recombination (e.g., penicillin and cephalosporin resistance in S pneumoniae) or during conjugative plasmid transfer (e.g., vancomycin resistance in S aureusEnterococcus faecalis). Like plasmids, transposons are extra-chromosomal, non-replicative DNA, lacking conjugative ability, able to transfer resistance genes among chromosomes or plasmids via a similar unique mobility.
Mutation of acquired genes involves DNA acquired from extra-chromosomes (foreign DNA) delivered to bacterial strains by plasmids, bacteriophages (bacteria-lysing viruses), naked sequences of DNA, or transposable elements (e.g., E coli production of TEM b-lactamase as first response to ampicillin; resistance in K pneumoniae mediated by b-lactamases derived by point mutation from the native TEM enzyme).6,7
Epidemiology
The expanding scope of antibiotic resistance and potential for new bacterial mechanisms has added to the strategies of mobile genetic elements and integron cassettes (e.g., NDM-1), which carry multiple resistance genes. The easy transfer facilitated by plasmids or rearrangement of large segments of DNA allow rapid spread of resistance among multiple strains of bacteria inevitable.
Transmission of NDM-1: The potential for outbreaks and the spread of disease has been exacerbated by increasing Gram-negative resistance to most available antibiotics (colistin and tigecycline excepted) and, more critically, by the paucity of drugs in development. Adding fuel to the fire is India’s history of easy-access to non-prescription antibiotics and the implications for selective resistance (e.g., the case of NDM-1).
In areas of Chennai and Haryana, India, NDM-1 has been widely spread throughout the environment. Concern mounted when the U.K. Department of Health issued a “National Resistance Alert 3” that NDM-1 had been introduced into the U.K. from India. Magnifying the prospect of international spread is travel to India and Pakistan, by both Europeans and Americans, for cost-effective elective (e.g., cosmetic) surgery.1
Failure of epidemiologic measures: In the past, infection-control guidelines for HA and CA infections (HAI, CAI) have proven effective in limiting the spread of infection. Failure of one or more techniques (e.g., effective hand-washing by healthcare workers), however, greatly increases transmission among patients. The spread of clonally-related organisms that colonize nares (e.g., nostrils, nasal passages), skin, and feces (e.g., MRSA, MDR Gram-negatives) results from a laxity of monitored hand washing and a decreased vigilance of isolation precautions.
Day-care centers, nursing homes, schools, military bases, and prisons all have become hot beds of CAI. Overuse of antibiotic agents, particularly in nursing homes, has created a pool of highly resistant organisms, easily transported to the hospital and back.
Last, contaminated food products are the end result of selective-resistant bacteria in animals treated with antibiotics as “growth promoters.” Salmonella, Campylobacter spp., E coli 0157 are typical Gram-negative bacteria with resistance tied to animal husbandry.6
New targets for therapy: Despite such measures as hospital antibiotic stewardship, which contributes greatly to preventing antibiotic overuse, the exploration of new approaches to the treatment and control of Gram-negative infection has been limited.
Human defense is based not only on enhanced infection-control measures but also in early tracking of resistance. Possible solutions include clinical laboratories’ routine use of molecular technology; results available for computerized surveillance; pursuit of new targets to deter bacterial resistance; [and] decreased dependence on the pharmaceutical industry.6
A study based on inhibiting a key component in the biosynthesis of an essential bacterial enzyme used to create nicotinamide adenine dinucleotide, or NAD, is an example of a new pursuit. Only target enzymes of bacteria used in the experiment (e.g., Franciscella tularencis, E coli, and Bacillus anthracis) were affected, not human enzymes. Application of this technology may be years away, but the potential for creating a new class of broad spectrum agents exists.8
Investigation into the tolerance of “persister cells” affords an alternative approach to the problem of chronic infection, which is even less treatable than acute infection. Persister cells are phenotypic variants of the bacterium wild type, tolerant to eradication by antibiotics (there is no effect on the MIC) because of their dormancy. An example is relapsing infection caused by biofilms. Certain compounds known as “prodrugs” (e.g., isoniazid or ethionamide used as anti-tuberculosis therapy, metronidazole for anaerobic bacteria) are capable of killing dormant cells. The presumption follows that a similar approach targeting bacterial enzymes may counteract the efflux mechanism common in Gram-negative resistance.9
Conclusion
The mechanisms of resistance that allow bacteria to survive and persist in the presence of antibiotics are well documented. For many years, Gram-positive bacteria (e.g., MRSA; vancomycin-resistant enterococci, or VRE; and Clostridium difficile) held public attention as primary causes of HAI and CAI. Isolates from severe infection (e.g., pneumonia, sepsis, surgical wound, skin, and urinary tract), however, invariably are Gram-negative bacteria. Since the 1990s, the appearance of ESBL-producing members of the Enterobacteriaceae have made MDR (i.e., resistance to at least three classes of antibiotics [e.g., b-lactams, carbapenems, fluoroquinolones, aminoglycosides, and so forth]), a household term.
MDR bacteria range from the ESBL-producing E coli and Klebsiella spp. to include the earlier carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii, as well as more recent carbapenem-resistant K pneumoniae and the MBL Enterobacteriaceae (NDM-1, IMP, VIM, and so forth). The endless production of enzymes by Gram-negative bacteria is solely responsible for out-of-control HAI and CAI; combined with mobile genetic elements that rapidly spread infection is the lack of new antibiotics in development, leaving their eradication to the guidelines of infection control.1
Poor coordination among healthcare systems, research and development, and the federal government continues to thwart the progress of targeted therapy, though reliance on pharmaceutical companies is no longer an option. Because antibiotic development is not economically feasible, searching elsewhere to solve the problem of bacterial resistance is critical to an ongoing global crisis. It bears repeating that infection-control guidelines are effective only when they are followed with scrupulous precision. Adherence to those well-established principles is documented to contain and prevent the spread of such infection as the resistant NDM-1, an organism capable of a proverbial death sentence.
Cynthia B. Schofield, MPH, MT(CAMT), is a microbiology technical supervisor (retired), VA San Diego Healthcare Systems, San Diego, CA.
References
- Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological and epidemiological study. Lancet Infect Dis. 2010;10(9):597-602.
- Yong D, Toleman MA, Giske CG, et al. Characterization of new metallo-b-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046-5054.
- Mollering RC Jr. NDM-1—a cause for worldwide concern. Perspective. NEJM. 2010;363(25):2377-2379.
- Centers for Disease Control and Prevention. Detection of Enterobacteriaceae isolates carrying metallo-beta-lactamase—United States 2010. MMWR. 2010;June 26:59(24):750.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 20th informational supplement M100-S20. Wayne, PA. 2010.
- Rice LB and Bonomo RA. Mechanisms of resistance to antibacterial agents. In: Murray PR, Jorgenson JH, Pfaller MA, et al, eds. Manual of Clinical Microbiology, 9th ed. Washington DC: American Society for Microbiology Press; 2007;(1):1114-1145.
- Opal SM, Medeiros AA. Molecular mechanisms of antibiotic resistance in bacteria. In: Mandell GL, Bennett JE and Dolin R, eds. Principles and Practice of Infectious Diseases, 6th ed. Philadelphia, PA; Elsievier Churchill, Livingstone Inc. 2005;(1):253-270.
- Sorci L, Yongping P, Eyobo Y, et al. Targeting NAD biosynthesis in bacterial pathogens: structure-based development of inhibitors in nicotinate mononucleotide adenyltransferase NadD. Chem Biol. 2009;16(8):849-861.
- Lewis K. Persister cells and the paradox of chronic infections. Microbe. 2010;5(10):429-437.