When asked which techniques and instrumentation we use in our laboratory to identify hemoglobin variants, our reply is high-performance liquid chromatography (HPLC), capillary zone electrophoresis (CZE), alkaline and acid electrophoresis, a CBC (complete blood count), calipers… and a telephone, when needed.1 Many times, the response is, “Isn’t that a little bit of overkill?”
The answer to that question is, simply, no. While not every patient specimen requires all of these technologies, the full force of clinical laboratory expertise and instrumentation is sometimes required to reliably identify a hemoglobin variant.
That’s because hemoglobin variant analysis is one of the most complex procedures carried out by clinical laboratories. Hemoglobin, the protein molecule in red blood cells that carries oxygen and carbon dioxide to and from the lungs to the rest of the body’s tissues, is made of four protein molecules (globulin chains).
A variant in the globin genes can cause a host of systemic disorders, from sickle cell and other forms of anemia to thalassemia. Reliable, precise identification and differentiation can have a significant impact on caring for patients with these and other blood disorders.
Take, for instance, the case of three patient specimens recently analyzed by our laboratory.
The three specimens were an Inkster, a G-Philadelphia, and a D-Los Angeles (D-Punjab). The HPLC retention times were very close to one another and at times truly could not be counted on to properly separate and identify these variants. Electrophoresis of all three variants (alkaline and acid gel) revealed an “S-like” migration on alkaline and an A migration on acid for all of the variant hemoglobins in question; therefore, it was not of any help in the differentiation. Ethnicity, obtained by telephone inquiry, was also of minimal assistance in the elucidation of the variants’ identities. Molecular and peptide analysis were available, but at significant expense.
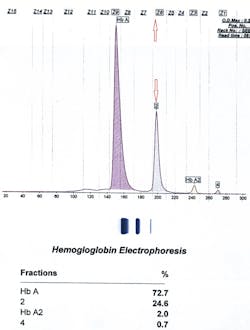
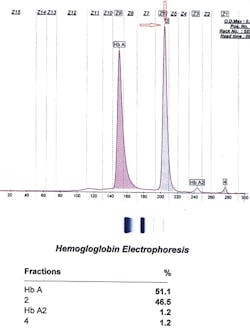
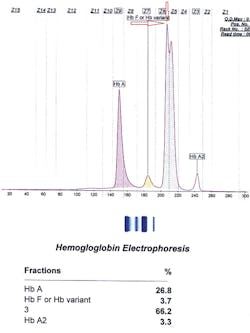
So we were left to ask, what other instruments could we use? How about CZE? It turned out that for these cases, CZE was the right choice. Below and on page 16 are the three tracings of hemoglobin Inkster, hemoglobin G-Philadelphia, and hemoglobin D-Punjab (in this case in combination with hemoglobin S).
Inkster (Figure 1) migrates in Zone 6 (Z6) closer to Z7. Hemoglobin G-Philadelphia (Figure 2) also migrates in Z6 closer to the middle of the zone, and hemoglobin D-Los Angeles (Figure 3) migrates also in Z6 but almost at the Z6/Z5 split point. These migrations are constant and duplicate from specimen to specimen and column to column, thus allowing this migrational difference to be a definitive interpretive factor in the identification and differentiation of these three hemoglobins. Inkster and G-Philadelphia, being benign alpha chain variant hemoglobins, also showed variant-specific delta chain variants, Inkster 2 and G-Philadelphia 2, shown as peak #4 in their tracings. D-Los Angeles, being a benign beta chain variant, does not show a separate D-LA 2 variant.
Identifying a hemoglobin variant sometimes requires employing a technology in a nonconventional manner. The example of hemoglobin Constant Spring comes to mind, often facetiously referred to in our laboratory as “Small Bump Disease.” Positive identification of the presence of Constant Spring is critical in that it can be a severe form of alpha thalassemia and a predictor of potential hemoglobin H disease in the future. Sometimes, however, the amount of Constant Spring is so miniscule that positive identification is very “iffy” at best. One technique to identify Constant Spring is alpha globin gene analysis for mutations and/or deletions. This molecular approach will provide definitive identification of any alpha globin mutation or deletion present, including Constant Spring. However, this analysis may at times take upwards of 10 days to two weeks to complete.
An alternative in these cases is to employ HPLC following a “concentration step”; that is, the instrument is forced to inject five times the amount of hemolysate onto the HPLC column. While not valid for peak percentage quantification, this approach does provide a significantly improved picture of the classic three-peak signature of hemoglobin Constant Spring. With a simple re-run of a patient’s specimen, we have been able to identify a potentially serious hemoglobin variant within a short time frame of about 10 minutes without resorting to more esoteric and expensive analyses.
As these cases of CZE differential migration and HPLC show, routine technologies, when creatively used, can provide high-quality results for the patient within a reasonable period of time. There is no such thing as using too much technology to perform reliable hemoglobin variant analysis. Laboratories require a full suite of tools and expertise to ensure they can deliver a result that is meaningful to the care of the patient.
Reference
- Burgess TE. Challenges in hemoglobinopathy testing. MLO. 2012; 44(2):18-20.