In the chapter of The Primer that appeared in the June 2013 issue of MLO (“Real-time PCR: the dyes that bind,” pages 34-37), we discussed the concept of real-time PCR and its advantages and disadvantages compared to more conventional gel resolved endpoint PCR. A major disadvantage of binding dye real-time methods, such as those employing SYBR Green I, was the lack of signal specificity. That is, in the event some non-specific products are created in the reaction, they will produce a signal which may be mistaken for a valid signal and lead to inappropriate conclusions and medical diagnosis. To improve on this, more advanced methods of real-time PCR with sequence-specific, probe-based fluorescent detectors have been developed. They are the topic of this chapter.
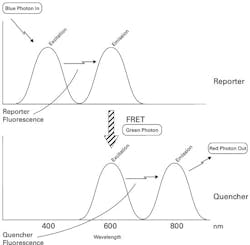
To understand the basis of these methods, we first have to consider an optical process known as Fluorescence Resonant Energy Transfer, or FRET. The reader will recall from the June Primer that fluorescence is the property whereby a dye molecule can absorb a photon of a defined wavelength range (its excitation spectrum) and rapidly re-emit this photon at a lower energy (and thus a shifted wavelength or color, in the dye’s emission spectrum). FRET occurs when two fluorophores are physically very close together, and the emission spectrum of one dye (known as the reporter) very closely matches the excitation spectrum of the second dye (referred to as the quencher). As diagrammed in Figure 1, under these circumstances exposure of the system to photons matching the reporter’s excitation spectrum get absorbed and re-emitted in that dye’s emission spectrum. When these emitted wavelengths are closely matched to the excitation spectrum of the quencher, and it is close to the reporter, this emitted photon is in effect “captured” by the quencher. A second fluorescence event then occurs, and the quencher re-emits a photon at a still longer wavelength. In the somewhat exaggerated example of Figure 1, a blue photon is absorbed by the reporter, a green photon passes from the reporter to the quencher, and a red photon is released by the quencher.
Note the requirement for very close physical proximity of the reporter and quencher for this process to occur. All of the probe-based methods of real-time PCR take advantage of this in some way by tying the process of amplification of a specific target sequence to either the co-localization of a reporter-quencher pair (causing FRET to start occurring) or the breaking apart of a closely associated reporter-quencher pair (causing FRET to stop occurring).
The first such method we’ll consider, and probably the one most commonly encountered in the molecular diagnostics laboratory, is the TaqMan chemistry made popular by ABI, now Life Technologies. In order to understand this method, we need to also know that some DNA polymerases (including common forms of Taq) have an additional enzymatic activity known as a 5′→3′ (or forward) exonuclease function. This activity allows the polymerase to “chew apart” single DNA strands it encounters that are base-paired to a template strand ahead of the polymerase as it makes a new strand. TaqMan chemistries take advantage of this and FRET together through the addition of short single-stranded synthetic DNA probes, labeled at one end with a reporter and at the other end with its paired quencher, to a PCR reaction. Critically, the sequence of the probe must be complementary to the sequence of the amplicon to be detected.
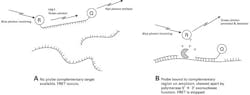
At the outset of a PCR reaction, there is little or no original template molecule for the TaqMan probe to hybridize to, and so it is free in the reaction solution. The real-time PCR machine illuminates the reaction tube with filtered photons matched to the reporter’s excitation wavelength, and uses a detector to “look” for photons at the reporter’s emission spectrum. Because of FRET, however, almost all photons released by the reporter dye are captured by the quencher dye, physically tethered at the other end of each probe molecule. In this situation (Figure 2A), little to no photon emission at the reporter’s emission spectrum can be detected. In the event, however, that correct amplicon (that is, one with an internal sequence complementary to the probe) is created during the PCR process, then at each PCR annealing step there is an available hybridization template for the probe molecules. After the probe anneals to this template, which in turn recruits a target specific PCR primer and DNA polymerase, extension by the polymerase occurs during the elongation phase. Reaching the annealed probe, the 5′→3′ exonuclease capable DNA polymerase breaks the probe apart, physically releasing the reporter dye from its quencher partner. Diffusion allows the reporter and quencher dyes originally conjugated to the now degraded probe molecule to separate from each other, and FRET can no longer occur with this probe molecule.
Now, when the reporter dye is illuminated at its excitation wavelength and emits photons at its emission wavelength, these photons are not captured by the quencher and escape the reaction tube to be detected by the real-time PCR instrument optics (Figure 2B). An increase in signal from the reporter molecule’s emission wavelength is therefore indirect proof that the PCR process has occurred in the reaction and that the product created contained the tested-for sequence, complementary to the TaqMan probe. In this way, TaqMan chemistry is doubly capable of selective identification of correct product, in that generation of product is shown, and sequence identity at the probe location is confirmed (akin to amplicon length confirmation by gel methods, or melt peak confirmation of amplicon identity as discussed with binding dye real-time methods).
By using spectrally selective filters on the real-time PCR instrument’s light source and detector, it is possible for this technique to multiplex based on the use of spectrally distinguishable reporter dyes. Many currently available instruments can separate up to four reporter channels, with some examples having six distinguishable channels. To support this, much research into FRET reporter and quencher dye pairs has been conducted, and the researcher or assay developer now has a wide spectrum of possible dye pairs to choose from. Further simplifying the detection process is the availability of “dark quenchers,” or quencher dyes which release their captured photons as heat or vibrational energy rather than as a photon. Doing so frees up visible spectral range for more reporter dyes to be used on an instrument.
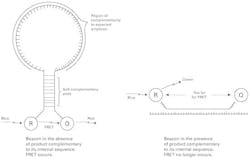
Alternate approaches to probe based real-time PCR include the use of molecular beacons, Scorpion probes, and hybridization probes. Molecular beacons (Figure 3) work by being long single stranded synthetic DNA oligonucleotides, with a reporter dye at one end and its FRET quencher pair at the other end. In this case, the probe length is quite long, the 5′ and 3′ ends of the oligonucleotide have short sections complementary to each other, and the long internal sequence of the scorpion probe is complementary to a sequence of the expected amplicon. In the absence of amplicon, the beacon’s ends will anneal to each other, forming a stem-loop structure. Formation of this stem brings the reporter and quencher dye pair extremely close to each other and allows for very efficient FRET to occur when no amplicon is present. If, however, amplicon is produced, the beacon will anneal to it via its internal section, thus moving the reporter and quencher pair apart from each other and breaking FRET. Similar to a TaqMan assay, the instrument is programmed to look for appearance of the reporter dye’s emitted photons as evidence for loss of FRET and thus presence of probe-complementary PCR product.
Scorpion probes build further on this idea by, in effect, using a molecular beacon which also has one of the target PCR primers added to one end of the beacon region. The molecule thus acts first as a primer, and if successful amplification occurs, the Scorpion will unfold to anneal its midsection to the complementary region of the product amplicon. As with a beacon, this breaks FRET. In contrast to TaqMan and beacon chemistries, Scorpion probes are unimolecular in their kinetics of action and thus offer potentially higher sensitivities than the other methods.
The hybridization probe approach to probe-based real-time PCR is to design two single-stranded probe molecules, with immediately adjacent regions of complementarity in the expected product amplicon. A reporter dye is conjugated to the end of one probe, and a matching FRET quencher is conjugated to the other probe in such a way that when both anneal to target, the reporter and quencher are located immediately next to one another. This method allows for “testing” a longer section of internal amplicon sequence for probe homology than the previous methods. Unlike the previous methods, this approach causes the occurrence of FRET rather than its breakage as evidence for successful, specific amplicon formation. This method has the added advantage that, similar to binding dye-based methods, a melt curve can be generated which permits still further certainty in product identity by allowing measurement of the denaturation temperature of the probes from their template.
Still further variations on the FRET probe theme for real-time PCR can include minor groove binding, or “MGB,” probes, which rely on additional molecular functions conjugated to the probe and increasing binding specificity, or the incorporation of non-standard nucleotide analogs such as peptide nucleic acids (PNAs) or locked nucleic acids (LNAs) to influence binding thermodynamics and specificity. While the details of these are beyond the scope of this chapter, their basic mode of action remains unchanged.
Probe-based real-time methods thus offer the advantages of binding dye real-time PCR (automation, decreased risk of amplicon contamination) but with increased specificity of result. When performed on instruments with multiple observation wavelengths of “channels,” rational selection of non-interacting FRET fluorophore pairs also allow probe-based real time methods to multiplex small numbers (2-5) of independent reactions within one tube; that is, multiple independent assays can be performed within one tube on a single sample. This is very useful for purposes such as a reaction internal control, used to assess that a sample does not inhibit a test PCR reaction being run simultaneously. Because of these advantages, probe-based methods are the mainstay of much of today’s clinical molecular diagnostics. All probe methods generate amplification curves similar to those discussed for dye binding real-time PCR discussed in the June 2013 installment of The Primer, and have CT values calculated in much the same way. (See the Figures in the June 2013 issue, pages 34-37.)
About the Author

John Brunstein, PhD
is a member of the MLO Editorial Advisory Board. He serves as President and Chief Science Officer for British Columbia-based PathoID, Inc., which provides consulting for development and validation of molecular assays.