Approximately 1.7 million hospitalized patients develop nosocomial infections annually while being treated for entirely unrelated conditions; more than 98,000 (one in 17) of these patients will die due to complications from acquired infections.1 Consequently, healthcare-associated infections are a major cause of morbidity and mortality around the world, posing an enormous financial and physical burden on the global healthcare system. Because of the increase in MRSA infection rates, hospitals have developed prevention and control measures to target patients at risk, many in conjunction with infection control. In addition, several states have passed or are considering legislation mandating the implementation of MRSA screening programs to reduce the risk to patients.
Methicillin-resistant Staphylococcus aureus (MRSA) was first reported in 1961, and the organism is now considered a major contributor of hospital-associated infections (HAI), resulting in tens of thousands of dollars annually in added per-patient costs. MRSA colonizes the anterior nares of approximately 1% of the population.2 Outbreaks occur when the microbe is transmitted through close contact, by contamination from environmental sources, or through improper hand hygiene practices by healthcare workers. Though MRSA is notable as a major pathogen in healthcare facilities, the organism can also cause illness in the community and is then commonly known as community-associated MRSA (CA-MRSA).
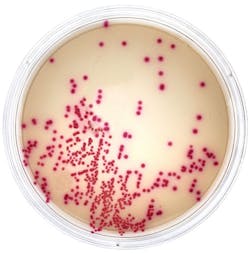
A selective and differential chromogenic medium for the qualitative detection of nasal colonization by MRSA
Approximately 10% of S. aureus strains in the U.S. are susceptible to penicillin, and many resistant strains remain susceptible to penicillinase-stable penicillins such as oxacillin and methicillin.3 A growing number of MRSA strains, however, have become resistant to all β-lactam agents, including cephalosporins and carbapenems. Moreover, due to complications surrounding immune-compromised patients in nursing homes, dialysis centers, ICUs, etc., HAI-MRSA strains may become resistant to other commonly used antimicrobials, making them much more difficult to treat.
Since the discovery of MRSA, its mechanisms of resistance have been well characterized. Resistance occurs when an isolate carries an altered penicillin-binding protein, PBP2a, mediated by the mecA gene.3 The mecA gene is part of the mobile genomic element Staphylococcal Chromosomal Cassette (SCCmec). There are four types of SCCmec, each with a varying degree of drug resistance. Community-associated MRSA carries SCCmec IV, which contains the mecA gene. SCCmec II and III are found in many nosocomial MRSA strains. These chromosomes harbor other antibiotic-resistance genes conferring resistance to aminoglycosides, tetracyclines, erythromycin, and clindamycin.
β-lactam antibiotics such as methicillin bind to proteins responsible for bacterial cell wall synthesis by a mechanism known as transpeptidation (peptidase enzymes, also known as penicillin binding proteins, PBP); thus, if PBP is altered, the β-lactam antibiotic cannot inhibit cell wall formation and the strain is rendered resistant. The Clinical Laboratory Standards Institute (CLSI) outlines screening and identification tests for detecting β-lactamase production, oxacillin resistance, and mecA-mediated oxacillin resistance to detect MRSA. These include the cefoxitin disk screen test, latex agglutination for PBP2a, or the MRSA Screen plate (Mueller Hinton Agar with 4% NaCl and 6µg/mL oxacillin).3,4
Less often described mechanisms that present a degree of non-susceptibility to methicillin are BORSA or MODSA, where BORSA hyper-produce β-lactamase and MODSA possess modified PBP. Neither of these modifications possesses the mecA gene and, therefore, may not produce traditional MRSA phenotypic profiles (i.e., full resistance). The minimum inhibitory concentration (MIC) of these strains to methicillin is usually in the intermediate range. Due to the scarcity of reports, the true clinical significance of these strains is still highly debated. However, it is felt that reporting them as MRSA is probably an overcall of resistance.
Detecting MRSA colonization in healthcare settings relies in part on an intricate balance among turnaround time, sensitivity, specificity, and cost. Culture using agar-based methods remains commonplace for MRSA surveillance; historically, these techniques account for more than 90% of the screening tests performed.5 However, a growing number of alternative methods, including broth-based enrichment, chromogenic media, rapid screening kits, molecular assays, and automated systems, are increasingly replacing traditional culture.3,5 In addition, isolating MRSA from screening swabs can often be a lengthy process due to the prevalence of competing flora; thus, finding a sensitive and specific method for rapid identification is critical for quick action.
Enrichment broth-based methods aid in increasing sensitivity and, in some cases, results are comparable with molecular-based tests.3,5 However, the increase in sensitivity comes at the expense of turnaround time. Selective agents such as methicillin, oxacillin, or cefoxitin are typically added to a 6.5% NaCl broth with indicator for early detection of MRSA, yet subculture is required for confirmatory testing.
A rapid, latex agglutination slide-card test to detect coagulase and/or protein A characteristics associated with S. aureus and MRSA
There is currently no defined standard for screening and isolation of MRSA using traditional agar-based methods. There are a variety of selective media available, usually containing NaCl and antimicrobials, including Mannitol Salt Agar with oxacillin and the MRSA Screen Plate (Mueller Hinton Agar with 4% NaCl and 6µg/mL oxacillin). Incubation and turnaround times vary using these formulations.
Over the last two decades, chromogenic culture media have increased in popularity, combining early screening and colony differentiation. These media traditionally incorporate a novel chromogen for differentiation in conjunction with the up-regulation of phosphatase activity: phosphatase is an enzyme present in all MRSA. To allow the medium to differentiate MRSA accurately, it also contains a combination of antibacterial compounds designed to inhibit the growth of a wide variety of competing flora. As a result, chromogenic media have an improved specificity and sensitivity compared to traditional agar-based formulations, and they have largely replaced standard culture due to a reduction in labor, ease of use, and increased speed.5
Molecular platforms for detecting MRSA rely largely on multiplexed PCR primers to detect genes specific to S. aureus and mecA methicillin resistance. Many utilize real-time polymerase chain reactions (PCR) for the amplification of MRSA DNA in conjunction with fluorogenic target-specific hybridization probes for the detection of the amplified DNA. Most FDA-cleared methods are essentially equivalent, providing increased sensitivities and specificities to enhance detection rates.5 Evidence shows that molecular methods increase speed and end-user convenience, but they are not designed to diagnose, guide, or monitor patient treatment, so concomitant cultures are necessary to recover organisms for epidemiological typing or for further susceptibility testing. Moreover, many users cite cost, which can be significant up front, and performance issues due to false-negatives (empty cassette variants) as reasons for converting back to standard culture methods.5
More recent advances in molecular tests utilize a fluorescein labeled, biotinylated DNA-RNA-DNA chimeric probe to provide enzyme specific action to detect the mecA gene. In these kits, target DNA negative results for the un-cleaved probe (mecA negative) demonstrate a colored end product; whereas, target DNA positive results permit cleavage of the labeled probe and prevent color formation.
Commercial agglutination kits for the detection of protein A and clumping factor are also readily available, but some strains of MRSA may have low levels of these proteins. These kits test for the presence of the mecA gene coding for methicillin resistance by detecting the PBP2' (penicillin binding protein 2') product using colored latex beads sensitized with monoclonal antibodies directed against PBP2'. Results are visualized against a white background where a positive reaction—agglutination (clumping)—indicates that the organism contains PBP2'.
Overall research shows that screening does reduce infection rates and institutional costs.2 Across patient groups, hospital screening generates an estimated savings of potentially thousands to tens of thousands of dollars monthly when compared to a lack of screening, not to mention the immeasurable value of saving lives.5 In addition, sensitivity analyses show that variations in transmission rates, conversion to infection, increases in prevalence, and hospital size are critical factors in determining the economic impact of screening programs. For all, though, turnaround time appears to be the common denominator in determining program efficacy.
Though there are improved test methods available for rapid screening and isolation of HAI-MRSA, recent literature suggests that work is needed in regard to the prevalence and virulence of healthcare-associated infections. Nurse-to-patient ratios, extended work hours, emotional exhaustion, and job-related burnout have all been linked to suboptimal care and increased infection rates.1 In addition, new research from Denmark shows that the loss of primary quorum-sensing-controlled virulence regulation in hospital-acquired staphylococcal isolates may lead to increased antibiotic resistance in these strains.6 In essence, selective pressure from antibiotic usage in hospital settings, including persistent low level concentrations of certain antibiotics commonly found in hospitalized patients, is driving a microclimate of evolutionary change likely to affect a multitude of infectious microorganisms. Thus, with hospitals under increasing financial pressure and a shrinking pool of effective antimicrobial chemotherapy options, it is vital that screening programs consider preventive action that not only offers better cost savings and more rapid results, but protects the efficacy of available antibiotics. If not, programs run the risk of merely identifying strains without a means to successfully combat them.
References
- Cimiotti JP, Aiken LH, Sloane DM, Wu ES. Nurse staffing, burnout, and health care-associated infection. Am J Infect Control. 2012;40:486-490.
- California Department of Public Health (CDPH). MRSA: methicillin-resistant Staphylococcus aureus. http://www.cdph.ca.gov/programs/hai/Pages/MRSAMethicillin-ResistantStaphylococcusaureus.aspx. Accessed November 13, 2012.
- Centers for Disease Control and Prevention (CDC). Laboratory detection of: oxacillin/methicillin-resistant Staphylococcus aureus. http://www.cdc.gov/HAI/settings/lab/lab_mrsa.html . Accessed November 13, 2012.
- Clinical and Laboratory Standards Institute (CLSI—formerly NCCLS). Performance standards for antimicrobial susceptibility testing; twenty second informational supplement. M100-S22. CLSI, Wayne, PA. 2012.
- Marlowe EM, Bankowski MJ. Conventional and molecular methods for detection of methicillin-resistant Staphylococcus aureus. Clin Microbio. 2011;49 (Suppl9):S53-S56.
- mBiosphere. For life in the hospital, staph drops its excess baggage. http://mbioblog.asm.org/mbiosphere/2012/11/for-life-in-the-hospital-staph-drops-its-excess-baggage.html. Accessed November 15, 2012.