Homeland security and bioterrorism applications:
Detection of bioweapon pathogens by microfluidic-based electrophoretic DNA analysis
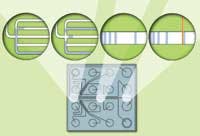
Figure 1. Schematic representation of the flow network architecture on the surface of a microfluidic LabChip. Electrophoretic separation and detection of DNA is conducted within a sieving polymer liquid that functions in a manner equivalent to slab gel separations.
Figure 2. Electrophoretic microfluidic chip-based
DNA analyzer (2100 Bioanalyzer, Agilent Technologies Deutschland
GmbH, Waldbronn, Germany). The instrument houses the prepped
microchip or LabChip, provides electrical power to run the
analysis, and incorporates an optical detection system. Signals
from the detector are sent to a PC equipped with application
specific software, which collects, processes, and reports the
results. To carry out an analysis, the chip is prepped with
reagents appropriate to the method (in this case, DNA 500) and
loaded into the instruments chip holder. Closing the cover
inserts electrodes into the chip wells and completes the
electrical circuit.
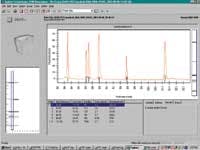
Figure 3. Identification of SARS Virus PCR product by the electrophoretic separation and detection of characteristic DNA fragments.*
Fragment lengths are determined from calibration curves, using a plot of standard ladder sizes vs. migration times. Ladders are part of the chip kit, which includes method-specific LabChips and pre-packaged chemistry for chip prepping. In addition to fragment size, the software also computes fragment concentration by ratioing peak area to an internal standard. Data can be presented as electropherograms, as PAGE-like images or in tabular format. An overlay feature enables graphical comparison of results from multiple wells, including the calibration ladder well. All analytical data, along with sample information, assay protocol, and annotated comments are archived in digital form and can be easily retrieved and distributed via intranets or the Internet.
*End-point PCR-amplified DNA obtained by RT-PCR amplification of whole-cell extract SARS virus RNA.
Table 1. Catalog of dangerous
biological agents/diseases
Category A:
The U.S. public health system and primary healthcare providers must be prepared to address various biological agents, including pathogens that are rarely seen in the United States. High-priority agents include organisms that pose a risk to national security because they
- can be easily disseminated or transmitted from person to person;
- result in high mortality rates and have the potential for major public health impact;
- might cause public panic and social disruption; and
- require special action for public health preparedness.
-
Anthrax (Bacillus anthracis)
-
Botulism (Clostridium botulinum toxin)
-
Plague (Yersinia pestis)
-
Smallpox (variola major)
-
Tularemia (Francisella tularensis)
-
Viral hemorrhagic fevers (filoviruses [e.g., Ebola, Marburg] and arenaviruses [e.g.,
Lassa, Machupo])
Category B:
Second highest priority agents include those that
-
are moderately easy to disseminate;
-
result in moderate morbidity rates and low mortality rates; and
-
require specific enhancements of CDCs diagnostic capacity and enhanced disease surveillance.
-
Brucellosis (Brucella species)
-
Epsilon toxin of Clostridium perfringens
-
Food safety threats (e.g., Salmonella species,
Escherichia coli O157:H7, Shigella) -
Glanders (Burkholderia mallei)
-
Melioidosis (Burkholderia
pseudomallei) -
Psittacosis (Chlamydia psittaci)
-
Q fever (Coxiella burnetii)
-
Ricin toxin from Ricinus communis (castor beans)
-
Staphylococcal enterotoxin B
-
Typhus fever (Rickettsia prowazekii)
-
Viral encephalitis (alphaviruses [e.g., Venezuelan equine encephalitis, eastern equine encephalitis, western equine encephalitis])
-
Water safety threats (e.g., Vibrio cholerae, Cryptosporidium
parvum)
Category C:
Third highest priority agents include emerging pathogens that could be engineered for mass dissemination in the future because of
-
availability;
-
ease of production and dissemination; and
-
potential for high morbidity and mortality rates and major health impact.
-
Emerging infectious diseases (such as Nipah virus and
hantavirus)
Source: U.S. Center for Disease Control (CDC) (https://emergency.cdc.gov/), December 30, 2003.
Today, we live with the knowledge that terrorism is a global threat. Since the events of September 11, 2001, we have become especially aware of our vulnerability to terrorism at home. Increasingly, we are learning to respond to this threat with countermeasures that include intelligence, increased security, and technology designed to detect weaponized chemical, biological, and radiological agents.
Biological weapons or bioweapons typically, pathogenic organisms and their toxic products constitute a particularly pernicious threat. They can be released into the air and into water systems or be otherwise disseminated (as was the case with anthrax spore contamination in the U.S. Postal System and at other sites). Unless suspicions are aroused and appropriate measures taken to sample contaminated environments, the presence of such agents is not usually confirmed until they produce symptoms in compromised individuals. If the agent is infectious, the affected individuals may become new vectors for spreading the disease unless they are identified, isolated, and treated (case in point, the recent SARS epidemic).
Therefore, the best approach for combating the bioterrorism threat requires implementing detection and containment measures and treating affected individuals as quickly as possible post-release. Since pathogens employed as bioweapons typically are highly toxic, detection methods should be rapid and unambiguous, and should be effective at very low analyte concentrations. High detection sensitivity and specificity are important, as false-negative or ambiguous results can have disastrous consequences for affected individuals who might go untreated or be wrongly treated. False-positives, on the other hand, might trigger inappropriate and/or wasteful mobilization of resources.
DNA-based microorganism identification
Since DNA can provide a unique signature for all organisms (certain viruses maintain their genomes in the form of RNA, which can be reverse transcribed into corresponding DNA sequences for analysis purposes), it is a logical focus for pathogen detection. At present, two approaches have been widely adopted for identifying organisms by characterizing their DNA: real-time polymerase chain reaction (PCR) and end-point PCR. Both techniques utilize specific sets of primers to amplify and detect DNA sequences unique to a given pathogen. In real-time PCR, detection of an organisms amplified DNA and therefore confirmation of that organisms presence is signaled by activation of a fluorescent dye reporter. End-point PCR, by contrast, does not use a fluorescent signal. Instead, the contents of each amplification well are subjected to electrophoresis and the migrating DNA fragments classified by molecular size as a function of the distance traveled. Each organism exhibits a characteristic DNA electrophoretic migration pattern by which it can be identified. In recent years, agarose and polyacrylamide gel electrophoresis have increasingly been replaced by a capillary version of this technique performed in microfluidic devices.
Microfluidic technology
An important innovation in the design of analytical platforms has been a reduction in size analogous to the downsizing of electronic circuits on microchips. Termed a chemical microchip, these devices contain interconnected fluid reservoirs and pathways fabricated on a glass surface that are sandwiched to form microchannels. Separation of molecules, such as DNA, RNA, or proteins by movement of analyte through these fluid-filled channels, is effected by electrokinetic forces (Figure 1). Advantages of microchip-based analytical methods include significant reduction in sample size and reagent usage, shorter analysis times, and improved resolution. Since microfluidic electrophoretic separations can be automated, gains are realized in both sensitivity and reproducibility as a result of the elimination of losses and errors from manual handling and transfer operations inherent in conventional gel methodologies.
Automation also extends to data handling. Both system parameters and analytical results are captured in digital form, thereby facilitating analysis, dissemination, and archiving of data, as well as precise replication of operating conditions. Microfluidic instrumentation and the corresponding methodologies for the electrophoretic separation and detection of DNA have been well established for several years. One system, 2100 Bioanalyzer, is capable of running 12 sample lanes in the microfluidic analog of a gel method (Figure 2).
Method comparison
A comparison of end-point PCR paired with microfluidic electrophoretic separation vs. real-time PCR with laser-scanning fluorescent detection reveals distinct advantages in the former approach. While both methods utilize 96-well plates for carrying out PCR amplification, real-time PCR is limited in the number of discrete amplifications that can be multiplexed within each sample well. Each amplification reaction requires a unique, optically resolvable fluorescent signal (nonoverlapping fluorescent emission), and this makes it impractical to run more than one or two target species per well. End-point PCR is not similarly constrained, since there is no detection required at the PCR amplification stage. As a result, it is possible to multiplex 10 to 20 discrete PCR amplifications within each well, gaining a corresponding increase in throughput and reduction in both the cost of consumables and instrument-use time.
Moreover, the ability to multiplex amplifications reduces the number of aliquots into which a sample must be subdivided to ensure comprehensive testing for all suspect organisms. This is not the case for real-time PCR for which the subdivision of samples into a large number of aliquots can significantly reduce test sensitivity in sample-limited situations. This distinction is important given the potentially serious consequences of a false-negative or ambiguous result resulting from insufficient sample size.
Detection of protein represents an alternative to DNA in pathogen identification. Generally, these enzyme-linked immunosorbent assays (ELISA) utilize specific antibody-antigen reactions to generate a confirmatory optical signal. Protein assays have many of the same limitations as real-time PCR, since unambiguous identification of a suspect organism usually requires running a battery of individual tests across all possible candidate species. Protein-based tests may also exhibit lower sensitivity than those based on DNA. They are also subject to concentration errors, and results can be confounded by temperature changes and other variations in test conditions.
Applications
The importance of both specificity and high throughput in pathogen detection cannot be overemphasized. Organisms can be packaged in ways that mask their identity and produce sets of symptoms that are not sufficiently characteristic early on for rapid and unambiguous identification. Since speed of analysis is essential for quickly formulating and implementing appropriate countermeasures, the ability to run tandem checks for multiple species eliminates much of the guesswork and associated delay in reaching a definitive result. Also, there is little ambiguity in identifying species by means of DNA electrophoresis, since the migration pattern correlates with species identity. Figure 3 is an electropherogram of SARS virus end-point DNA determination. SARS primers published by the World Health Organization
(WHO)1 were used to amplify DNA obtained, in turn, by RT-PCR amplification of whole-cell extract SARS virus
RNA.2 A multiplex PCR method is currently under development for screening the entire CDC A-List Pathogens (Table 1).
Conclusion
Microfluidic chip-based electrophoretic analyzers and the corresponding methods for DNA separation and identification are
well-established.3,4,5 The challenge of adapting these techniques for pathogen identification has been met and is now being elaborated for the most important bioweapon pathogens. These results demonstrate that we now have the analytical capability to address the critical needs of pathogen detection in support of homeland security/counter terrorism efforts. The logical next step would be better integration of the individual operations of DNA isolation, amplification, and microfluidic-based analysis. Looking somewhat further out, we can anticipate the development of an end-to-end detection system utilizing microfluidic technology to move the entire pathogen analysis stream into a chip-based analytical system.
Thomas Gluodenis is homeland security marketing manager, and Scott Harrison is agbiotech industry marketing manager for Agilents Life Science and Chemical Analysis Group.
References
- WHO. PCR primers for SARS developed by WHO Network Laboratories. Available at:
www.who.int/csr/sars/primers/en/. Accessed April 17, 2003. - Bellini WJ, PhD, Chief, Measles Virus Section, Respiratory and Enteric Viruses Branch, DVRD/NCID/CDC. Acknowledgement. Whole cell extract SARS RNA.
- Mues MB, Koehler CI, Odenthal M. Microfluidic analysis of multiplex PCR products for the genotyping of Helicobacter pylori. 2003; 5989-0078EN.
www.chem.agilent.com/temp/rad657F6/00044782.pdf. Published October 1, 2003. Accessed December 5, 2003. - White S, den Dunnen J. Rapid detection of genomic duplications and deletions using the Agilent 2100 bioanalyzer, 2003;5989-0192EN.
www.chem.agilent.com/temp/radC978C/00045036.pdf. Published October 1, 2003. Accessed December 5, 2003. - Valer M. Measuring multiple apoptosis parameters with the Agilent 2100 bioanalyzer, A simplified and fast protocol for the analysis of DNA fragmentation during apoptosis. 2003;5988-8028EN.
www.chem.agilent.com/temp/radE7B96/00038171.pdf. Published October 1, 2002. Accessed December 5, 2003.
February 2004: Vol. 36, No. 2