Automating tissue dissection key to personalized medicine
Globally, the incidence of cancer continues to rise, with an anticipated 27.5 million new cases occurring each year by 2040. The American Cancer Society estimates that the United States will have 1.9 million new cancer cases this year, with 609,360 deaths.1
Advances in molecular biology have led to better understanding of molecular mechanisms behind the disease and laid the fundaments for targeted therapies addressing specific driving mechanisms behind each individual cancer (personalized therapy).2-4 With advanced stage cancers where surgery is no longer an option, targeted therapies are often the last line of defense.
Molecular diagnostics holds the key to stratifying patients for the best therapy, and next-generation sequencing (NGS) is becoming available for routine clinical diagnostic laboratories. However, sample selection for molecular analysis presents numerous challenges for the pathologist and the laboratory.5 The sensitivity and specificity of the emerging molecular diagnostic tests depend on the quality of the sample, in particular the number and purity of tumor cells in the sample.
The National Comprehensive Cancer Network (NCCN) addressed some limitations of current tissue selection and dissection methods in its recently issued 2023 guidelines for molecular and biomarker analysis for advanced cancers. These included specific guidance when the laboratory had insufficient tissue to review due to the clinician seeking to minimize the invasive procedure for the patient.6 Tests need a minimum amount and percentage of tumor cells in the sample; but at the moment, this is not really measured. Pathologists have to make an estimate based on their experience.
Novel technology solves missing link
It is here we see the potential flaw in the expansion of personalized therapies.7 While other processes have been automated and integrated into a laboratory’s workflow, there has been little innovation in the area of sample selection and dissection. These steps remain time-consuming manual processes, relying on subjective analysis, prone to ‘misestimation’ with limited precision and the risk of contamination.8 Variability, poor quality control, and lack of traceability are other pitfalls.
Novel technology was needed that would address this missing link. But any new solution cannot just offer improved quality. It must address the challenges set by higher performance targets, over-stretched resources, and limited staff availability. Hands-on time must therefore significantly be reduced, and less experienced staff should be able to safely use it.
Once the process is set in motion, staff must be able to walk away and direct their attention to other, added-value tasks. Further, it should be flexible enough to integrate into the existing routine workflow. Compare this to what is still taking place in today’s histopathology laboratory. Current practice is for the experienced histopathologist to have to mark the regions of interest (ROIs) by hand on hematoxylin and eosin (H&E)-stained slides, using subjective analysis to determine the boundaries of the tumor area. This manual process becomes increasingly difficult with smaller regions of interest.
Need to address vulnerabilities
For an automated system to add full value, it must offer an accuracy of ≤0.1 mm — being able to run uninterrupted for around two hours would be optimal. The number of slides handled in an hour, along with the ability to add more cases without interrupting the operation and freely selecting the number of dissection slides per case are all important requirements. Pathologists also look for time-saving features such as automatically generated quality control (QC) reports containing pre- and post-dissection images and quality metrics for full traceability.
These requirements were reinforced by findings from a recent global study involving clinicians and laboratory scientists (commissioned by Xyall BV). They commented on the vulnerabilities of manual detection and the need for faster and more accurate tissue dissection so that precision medicine could become a reality. The findings also pinpointed how accuracy and cross-contamination were two of the biggest concerns of current practice in molecular diagnostic laboratories worldwide. Quality control was also an area of concern.
The respondents warned that current practices of manual dissection of tissue were seen as a hindrance to efficiency and turnaround time, with around a third directly calling for this to be automated. Lack of automation in tissue dissection was seen as ‘a glaring gap’ in the otherwise automated laboratory workflow process.
Pioneering US laboratory
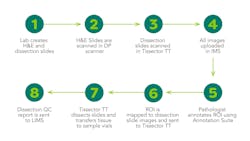
This has now become a reality with the United States leading the way globally, where one of the world’s leading research institutes has just installed the first compact tabletop platform (Tissector Table Top, Xyall BV). This automates and integrates the entire process (See Figure 1).
With an automated system, pathologists can make a selection of the region of interest by assessing digital whole-slide images of the sample. The annotations of the regions of interest are then interactively created and automatically transferred to dissection slide images. These steps are digitally captured and can be reassessed at any stage. Automation also makes it possible to annotate the ROI remotely, from wherever the pathologist is working in the hospital. Another benefit of having full traceability is that results can be quickly accessed for further review by a molecular tumor board.
This is in direct contrast to current practice, where histopathologists review the tumor tissue under a microscope to identify ROIs. Using their experience, they make an educated guess on the tumor cell percentage, marking the outline of these ROIs on an H&E-stained slide. The technician then manually transfers the annotated area from the reference slide to a number of unstained dissection slides, matching these by visual judgement. The dissection operation itself is often done with a scalpel, with the technician manually following the annotation as accurately as possible. Novel technology would replace these processes, automatically collecting tissue material, operating continuously, with the risk of cross-contamination minimized by the automatic disposal of the scraping tool. With no need for liquids, compliance with all molecular sample preparation protocols is warranted.
What the industry needed was an automated system using image registration algorithms to automatically map the marked ROI slides to the dissection slides. The coordinates of the transferred annotations can then automatically guide the robotic dissection arm so that it removes the material from the region of interest with high precision. The same device then collects and transfers the material to the sample tube, automatically replacing the scraping head to minimize the risk of cross-contamination. It is also easier to comply with protocols for molecular sample preparation when no liquid is used.
The evidence for automation
There are several studies giving evidence of the diagnostic benefits of automated tissue dissection. In a study published in Cancer Genetics, the authors performed and compared digitally guided dissection with traditional manual dissection in a series of pancreatic adenocarcinoma specimens, to compare the effectiveness of both methods.9 The researchers found that the KRAS mutant allele fraction and estimated neoplastic cell fraction were significantly higher in samples obtained from digitally guided dissection. In 7 out of 32 (22%) of the samples, a detectable mutation was found only with the digitally guided dissection.
Further, a 2022 study published in Modern Pathology highlighted deficiencies in the way ‘misestimation may cause tissue waste and increased laboratory costs.’10 The same study also indicated the benefit of combining automated tissue dissection with artificial intelligence (AI) for accurately determining the parameters of ROIs whatever the clinical analysis.
The study noted the particular challenges pathologists face to achieve minimum DNA input requirements for NGS. Without any automation solution, they currently have to visually estimate the dissection areas and slide count decisions, while taking care not to recommend excessive dissection. Tissue stewardship guidelines help them to protect tumor tissue in case it is needed for further molecular tests — but they still have to rely on subjective interpretation.
It further points out that ‘using manual dissection techniques is difficult, and thus, there is an increasing need to optimize tissue extraction procedures as NGS becomes more relevant in clinical practice.’ An algorithm in combination with an automated system would enable more tissue to be preserved and avoid excessive dissection. An obvious development from this would be the addition of software that would flag up to the pathologist if an ROI needed further review.
Delivering greater workflow efficiency
For hospital-based histopathology, the digitization of whole-slide imaging is already transforming workflow with AI-based algorithms becoming available for assessing tissue morphologies, classifying cancer subtypes and segmenting the tumor area. Potentially, AI-based algorithms could also be used in the selection of ROIs. However, this remained impractical while sample scraping was carried out manually.
Histopathology has benefited hugely from the digitization of whole-slide imaging, allowing remote discussions (telepathology), image archiving, improved storage and retrieval, comparison of areas on different slides, and direct slide annotation. This delivers increased efficiency, faster turnaround time, high-throughput sample processing, and increased reproducibility. It also makes best use of scarce and valuable staff resources.11 The development of a solution to the missing link — automated tissue dissection — offers a compelling way forward, able to integrate into a laboratory’s automated workflow and deliver robust, standardized molecular diagnostic testing for laboratories of all sizes.
References
1. Cancer facts & figures 2023. Cancer.org. Accessed November 3, 2023. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2023-cancer-facts-figures.html.
2. Beaubier N, Bontrager M, Huether R, et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat Biotechnol. 2019;37(11):1351–1360. doi:10.1038/s41587-019-0259-z. (https://www.nature.com/articles/s41587-019-0259-z).
3. Wagle, P, Nikolić, M, Frommolt P. QuickNGS elevates next-generation sequencing data analysis to a new level of automation. BMC Genomics. 2015;16(1):487. doi:10.1186/s12864-015-1695-x. (https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-015-1695-x).
4. Hess JF, Koh TA, Kotrová M, et al. Library preparation for next generation sequencing: a review of automation strategies. Biotechnol Adv. 2020;41:107537. doi:10.1016/j.biotechadv.2020.107537. (https://www.sciencedirect.com/science/article/pii/S0734975020300343?via%3Dihub).
5. Al-Kateba H, Nguyena T T, Steger-May K, Pfeifer J D. Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (NGS). Mol Oncol. 2015;9(9):1737-43. doi:10.1016/j.molonc.2015.05.004.
6. NCCN guidelines. NCCN. Accessed November 3, 2023. https://www.nccn.org/guidelines/nccn-guidelines.
7. Schwartzberg L, Kim ES, Liu D, Schrag D. Precision oncology: who, how, what, when, and when not? Am Soc Clin Oncol Educ Book. 2018;37:160–169. doi:10.1200/EDBK_174176. (https://ascopubs.org/doi/10.1200/EDBK_174176?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed)
8. Smits AJ, Kummer JA, de Bruin PC, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol. 2014;27(2):168–174. doi:10.1038/modpathol.2013.134. (https://www.modernpathology.org/article/S0893-3952(22)01806-3/fulltext).
9. Geiersbach K, Adey N, Welker N, et al. Digitally guided microdissection aids somatic mutation detection in difficult to dissect tumors. Cancer Genet. 2016;209(1–2):42–49.
10. Osinski BO, Taieb AB, Ho I, et al. Artificial intelligence-augmented histopathologic review using image analysis to optimize DNA yield from formalin-fixed paraffin-embedded slides. Mod Pathol. 2022;35(12):1791–1803. doi:10.1038/s41379-022-01161-0. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9532237/).
11. Evans AJ, Vajpeyi R, Henry M, Chetty R. Establishment of a remote diagnostic histopathology service using whole slide imaging (digital pathology). J Clin Pathol. 2021;74(7):421-424. doi:10.1136/jclinpath-2020-206762.
About the Author

Reinhold Wimberger-Friedl, PhD
is Xyall’s Chief Technology Officer. He has a PhD in mechanical engineering and a master’s in chemical engineering. Reinhold has been at the cutting edge of research and development in the medical diagnostics industry for many years, co-authoring more than 100 patent applications. Previously a principal scientist at Philips, he worked on innovations in digital and molecular pathology as well as new diagnostic technologies, like MiniCare (Siemens Healthineers) and Idylla (Biocartis).