T cells mount attacks against many SARS-CoV-2 targets
The new research, published in Cell Report Medicine, is the most detailed analysis so far of which proteins on SARS-CoV-2 stimulate the strongest responses from the immune system’s “helper” CD4+ T cells and “killer” CD8+ T cells, according to LJI.
The immune system is very flexible. By re-scrambling genetic material, it can make T cells that respond to a huge range of targets, or epitopes, on a pathogen. Some T cell responses will be stronger against some epitopes than others. Researchers call the targets that prompt a strong immune cells response “immunodominant.”
For the new study, the researchers examined T cells from 100 people who had recovered from SARS-CoV-2 infection. They then took a close look at the genetic sequence of the virus to separate the potential epitopes from the epitopes that these T cells would actually recognize.
Their analysis revealed that not all parts of the virus induce the same strong immune response in everyone. In fact, T cells can recognize dozens of epitopes on SARS-CoV-2, and these immunodominant sites also change from person to person. On average, each study participant had the ability to recognize about 17 CD8+ T cells epitopes and 19 CD4+ T cell epitopes.
The new study shows that while the immune system often mounts a strong response against a particular site on the virus’s “spike” protein called the receptor binding domain, this region is actually not as good at inducing a strong response from CD4+ helper T cells.
Without a strong CD4+ T cell response, however, people may be slow to mount the kind of neutralizing immune response that quickly wipes out the virus.
Among the many epitopes they uncovered, the researchers identified several additional epitopes on the SARS-CoV-2 spike protein.
Researchers from the University of California, San Diego, Australia’s Murdoch University collaborated on the project.
Mount Sinai identifies three molecular subtypes of Alzheimer’s disease
Researchers at the Icahn School of Medicine at Mount Sinai have identified three major molecular subtypes of Alzheimer’s disease (AD) using data from RNA sequencing and published their findings in Science Advances, according to a press release from the medical school.
RNA is a genetic molecule similar to DNA that encodes the instructions for making proteins, while RNA sequencing is a technology that reveals the presence and quantity of RNA in a biological sample, such as a brain slice.
There is growing evidence that disease progression and responses to interventions differ significantly among Alzheimer’s patients. Some patients have slow cognitive decline, while others decline rapidly; some have significant memory loss and an inability to remember new information, while others do not; and some patients experience psychosis and/or depression associated with AD, while others do not.
The research team analyzed RNA-sequencing data of more than 1,500 samples across five brain regions from hundreds of deceased patients with AD and normal controls, and identified three major molecular subtypes of AD, independent of age and disease stage.
These subtypes correspond to different combinations of multiple dysregulated biological pathways leading to brain degeneration. Tau neurofibrillary tangle and amyloid-beta plaque, two neuropathological hallmarks of AD, are significantly increased only in certain subtypes.
Many recent studies have shown that an elevated immune response may help cause Alzheimer’s. However, more than half of AD brains do not show increased immune response compared to normal healthy brains. The analysis further revealed subtype-specific molecular drivers in AD progression. The research also identified the correspondence between these molecular subtypes and the existing AD animal models used for mechanistic studies and for testing candidate therapeutics.
Although the subtyping described by the researchers was performed postmortem using the patients’ brain tissue, the researchers said that if the findings were validated by future studies, they could lead to the identification in living patients of biomarkers and clinical features associated with these molecular subtypes and earlier diagnosis and intervention.
Common HIV drugs may help prevent leading cause of vision loss
Scientists have identified a group of drugs that may help stop a leading cause of vision loss after making an unexpected discovery that overturns a fundamental belief about DNA, according to a University of Virginia news release.
The drugs, known as nucleoside reverse transcriptase inhibitors, or NRTIs, are commonly used to treat HIV. The new discovery suggests that they may be useful against dry macular degeneration as well, even though a virus does not cause that sight-stealing condition.
A review of four different health insurance databases suggests that people taking these drugs have a significantly reduced risk of developing dry macular degeneration.
The new discovery comes from researchers at UVA, the Salk Institute for Biological Studies, and collaborators around the world. The work rewrites scientists’ understanding of DNA, revealing that it can be manufactured in the cytoplasm of our cells, outside the cell nucleus that is home to our genetic material.
The buildup of a certain type of DNA in the cytoplasm, Alu, contributes to macular degeneration, the researchers found. This buildup appears to kill off an important layer of cells that nourishes the retina’s visual cells.
Based on this discovery, the researchers decided to look at drugs that block the production of this DNA, to see if they might help prevent vision loss. They found that people taking NRTIs were almost 40 percent less likely to develop dry macular degeneration.
Gestational diabetes can be tip-off to future increased risk for heart disease
Women with a history of gestational diabetes are at increased risk for heart artery calcification, a marker of increased risk for heart disease, throughout their childbearing years and into mid-life, even if they currently have normal blood sugar levels, a newly released study from Kaiser Permanente Division of Research shows.
The study, published in Circulation, is the first to look at heart disease risk in relation to changes in blood sugar levels in women who developed gestational diabetes and those who did not over a 25-year period, Kaiser Permanente Division of Research said.
The study found a 2-fold higher risk for heart artery calcification in women who had gestational diabetes and normal blood sugar, compared with women who did not have gestational diabetes and had normal blood sugar levels. The 2-fold higher risk was present in women with a history of gestational diabetes who many years later had blood sugar levels classified as normal, pre-diabetes, or type 2 diabetes.
Erica P. Gunderson, PhD, MS, MPH, Senior Research Scientist at the Kaiser Permanente Northern California Division of Research, led the study.
Yearly, in the United States, about 250,000 pregnancies develop diabetes.
Because gestational diabetes greatly increases a woman’s lifelong risk for type 2 diabetes, the American Diabetes Association recommends that all women who develop gestational diabetes have a glucose tolerance test every 1 to 3 years. American Heart Association guidelines underscore that a history of gestational diabetes is important to consider in evaluating individual risk for atherosclerotic heart disease.
The new findings suggest physicians should monitor factors for heart disease in all women who develop gestational diabetes, before they develop prediabetes or diabetes. Women with a history of gestational diabetes may benefit from healthy lifestyle choices and weight loss.
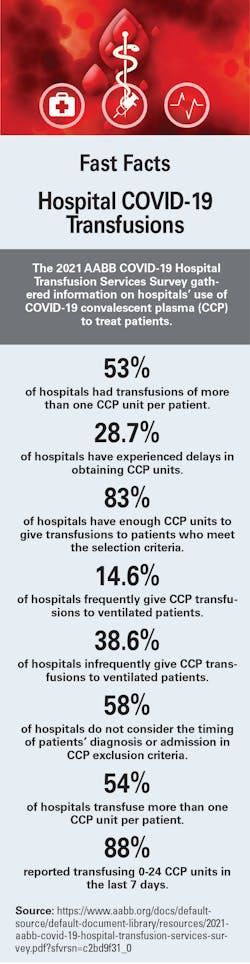
Subset of COVID-19 patients have increased bleeding riskResearch has found that patients with COVID-19 are prone to serious blood clotting. However, a new study from Michigan Medicine found that aside from this heightened clotting risk, some COVID-19 patients have an unbalanced ability to break down clots as well, which is linked to a potential clinical biomarker seen in later stages of the disease, according to a news release. The study was published in Scientific Reports and led by senior author Daniel Lawrence, PhD, Professor of Basic Research in Cardiovascular Medicine at Michigan Medicine. The abnormal process of breaking down clots can contribute to a high bleeding risk, raising concerns about the current practice of giving COVID-19 patients high-dose anticoagulants throughout the duration of their disease course. The study from Michigan Medicine included 118 COVID-19 patients and 30 healthy controls. In the COVID-19 patients, the team expected to see high levels of plasminogen activator-inhibitor-1, a molecule associated with stabilizing blood clots. However, they did not expect high levels of tissue-type plasminogen activator, the molecule responsible for removing the clots. Almost half of the study’s patients were supported by a ventilator and a quarter breathed just room air. Compared with the patients breathing room air, patients that required supplemental oxygen had significantly higher levels of plasminogen activator-inhibitor-1, but not of tissue-type plasminogen activator. High levels of both tissue-type plasminogen activator (tPA) and plasminogen activator-inhibitor-1 (PAI-1) were associated with worse lung function, but high tPA was independently correlated with mortality. The levels of either molecule can increase independently of one another, but the research found a change in one can have consequences on the other. The team asked whether COVID-19 plasma with the highest tPA levels might correlate with an enhanced, spontaneous breaking down of clots, as compared with low tPA COVID-19 plasma or control plasma. After assessing 10 COVID-19 plasma samples with high tPA, 10 COVID-19 samples with low tPA, and 10 healthy control plasma samples, it was clear the high-tPA COVID-19 samples were found to significantly enhance spontaneous clot breakdown compared to the other two groups. The researchers said this means that high tPA may be a biomarker for high bleeding risk and poorer outcomes in COVID-19. The researchers suspect the source of these high levels of tPA in COVID-19 patients, and the subsequent clotting issues, is because of damage to endothelial cells, which are cells that line blood vessels. If badly damaged, the blood vessels can actually break and cause bleeding. The theory is that a hallmark symptom of COVID-19 ARDS, when fluid builds up in the lungs and causes trouble breathing and low oxygen levels in the blood, may trigger endothelial cell activation, which consequently promotes the release of tPA. |