Phenotypic analysis of bacterial virulence using electric cell-substrate impedance sensing
The clinical microbiology laboratory is a fundamental element of patient care.1 Typically, clinical material is obtained by a healthcare professional and presented to the laboratory for analysis.2 This material is then processed by the laboratory staff, resulting in the derivation of a bacterial isolate. This isolate is then interrogated by genotypic and/or phenotypic methods to determine genus, species, and antimicrobial resistance properties. It is then either archived or discarded, depending upon laboratory protocol. Typically, no attempt is made to characterize the isolate beyond antimicrobial discrimination, and no attempt is made to estimate virulence.3
Since bacterial infections are often difficult to treat and the application of inadequate treatment regimens cultivates the development of resistant phenotypes, however, isolate characterization is an essential prerequisite to treatment.4,5 We propose that the introduction of technology capable of evaluating the ability of a bacterial isolate to alter host-cell barrier function can serve as a supplement to antimicrobial resistance studies. Furthermore, we propose that a novel technology known as electric cell-substrate impedance sensing (ECIS) can function as an autonomous, scalable, rapid, and effective means of evaluating the impact of individual bacterial isolates on host-cell barrier function.6,7
Damage of host cells as an estimate of virulence
Bacterial pathogens damage host cells by multiple mechanisms.8,9 The damage can have a wide variety of outcomes depending on the site of infection, the degree of infection, and the cell types involved. One consequence of cell damage is cell death (apoptosis). It has been demonstrated that certain strains of cytotoxic Staphylococcus aureus are capable of inducing apoptosis in host endothelial cells.10 Several genera of gram-negative bacteria have been shown to exhibit similar activity in epithelial cells.11,12 It is tempting to speculate that bacteria-induced apoptosis is an adaptive phenomenon that serves to facilitate bacterial spread and reproduction.13
Since endothelial membranes line the vasculature and epithelial cells line the gastrointestinal tract and mucosal surfaces, bacteria-induced destruction of these cells can result in the loss of tissue integrity.14 The loss of tissue integrity can potentially allow the passage of bacterial pathogens across tissue surfaces from one compartment to another and result in an enhanced and disseminated infection.15 It is interesting to note that bacterial pathogens have been identified that are capable of modulating the tight junctions that are responsible for the maintenance of host membrane integrity.16
The destruction of host cells and the modulation of host membrane integrity has been found to be dependent on the secretion of specific proteins from the pathogen. Hence, in order to damage host cells, it appears that the pathogen needs to be metabolically active and capable of synthesizing the required proteins, transporting those proteins across the cell membrane, and shuttling them across the cell wall.17
Given these details, it is reasonable to suspect that the relative capacity of a bacterial isolate to degrade the barrier function of cell monolayers can serve as an estimate of bacterial virulence. Furthermore, it is reasonable to suspect that the degree to which an antimicrobial agent is capable of attenuating or inhibiting the ability of a pathogen to degrade barrier function can serve as an indication of the ability of the agent to attenuate or inhibit overall virulence.
Evaluating damage via microscopy
In order to assay the impact of bacterial co-culture on eukaryotic cells, several methods can be utilized to measure cell damage. Chromogenic and fluorescent staining methods are among the most efficient and economic methods available. For example, trypan blue is a compound that is not capable of crossing an intact eukaryotic cell membrane. It can, however, cross damaged membranes, and although it is routinely used in the cell biology laboratory to discriminate between live and dead cells, it can also be used to identify cells with damaged membranes.18
A more detailed methodology has been described that uses calcein-AM to monitor extracellular enzyme activity and ethidium homodimer-1 to measure cell membrane integrity.19 Calcein-AM fluoresces green upon reduction by host cell enzymes; ethidium homodimer-1 fluoresces red. Metabolically active cells with an intact cell membrane fluoresce green, dead cells with a ruptured cell membrane fluoresce red, and metabolically active cells with damaged cell membranes fluoresce yellow. Calcein-AM/ethidium homodimer-1 assays for cell viability determination are commercially available and yield impressive results. A typical experiment for demonstrating the effect of bacterial co-culture on eukaryotic cell viability would involve culturing a specific eukaryotic cell type such as endothelial cells. However, staining methods are typically end-point assays and are not capable of measuring cell viability or metabolic activity in real-time.20 In addition, these methods can be laborious and time-consuming, particularly when numerous controls and treatment regimens need to be interrogated.
ECIS as a means of monitoring damage
Electric cell-substrate impedance sensing (ECIS) is an emerging technology capable of interrogating the morphology of eukaryotic cells in real-time.21 This is achieved by growing the cells on gold electrodes that coat the bottom of a modified cell culture dish. As the cells grow to cover the surface of the electrode, they impede the flow of an externally applied alternating current through the culture media. As this occurs, an electronic monitoring system measures changes in impedance as a function of time. The data obtained by monitoring impedance changes can be used to model cell barrier function, membrane permeability, and cell metabolism.22
Although ECIS technology has not been previously applied in the clinical microbiology laboratory, it has been shown to be effective in monitoring the cytotoxic effects of viruses on eukaryotic cells, and unpublished experiments in our laboratory have shown that it is capable of detecting bacterial induced cell damage.6 ECIS technology has several advantages over microscopy for monitoring the effects of bacteria on host cells. The most important advantage is that ECIS is a real-time continuous impedance monitoring system.7 We hypothesize that real-time monitoring of bacterial-induced cell damage is superior to end-point analysis and that this superiority is due to the ability of this system to determine the kinetics of cell monolayer barrier degradation at very early stages and ending in cell death. The resulting data set can potentially be used to model the response of a given cell type to a given bacterial strain, determine differences in response between various strains, and evaluate the impact of antibiotics on this response.
ECIS in the microbiology lab
Typically, clinical microbiology laboratories provide healthcare providers with two pieces of information: the identity of the pathogen, and its antimicrobial resistance properties.23 ECIS technology has the potential to augment the data provided by the laboratory by serving as an adjunct to traditional antimicrobial susceptibility testing and by allowing the direct phenotypic measurement of an indicator of bacterial virulence. When a clinical sample enters the lab, it is usually processed and plated on various media in order to grow and isolate the etiologic agent.
Once the pathogen is isolated, it can then be identified by morphology, biochemical properties, or molecular characteristics. Antimicrobial sensitivity studies are usually performed in parallel with pathogen identification. Furthermore, antimicrobial resistance profiles based upon growth profiles of the pathogen in the presence of antibiotics are often used to guide treatment.24
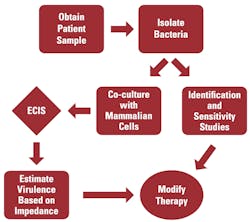
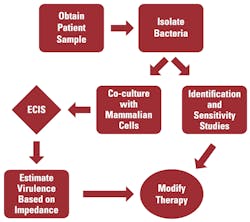
The addition of ECIS to the clinical laboratory should not significantly alter the current workflow. ECIS can be performed in parallel with routine identification and sensitivity studies utilizing only a small portion of the clinical isolate. For example, a single colony of the isolate can be grown in liquid medium until a predetermined optical density is obtained. The resulting bacterial suspension can then be used to inoculate a set of confluent cells that are growing on gold-electrodes. Cell lines can be selected based upon their ability to best model the current clinical picture. (For example, uroepithelial cells can be used to model urinary tract infections and vascular endothelial cells can be used to model vascular tissue.) An un-inoculated culture will serve as a negative control, and a highly cytotoxic bacterial strain will be used as a positive control. Impedance will then be measured using the ECIS system for a pre-determined period of time, and any loss of impedance in the inoculated culture, within a specific time frame, will be interpreted as a loss of barrier function resulting from bacterial cytotoxic activity. If the pathogen rapidly degrades barrier function, clinicians will be notified so that they are aware that the patient has a possibly invasive, cytotoxic strain of bacteria. ECIS data can also be utilized to develop a strategy for antibacterial therapy and to determine whether aggressive intervention is required (Figure 1).
Further, patient-derived bacteria and cells can be co-cultured in the presence of various antibiotics to determine whether antibiotic treatment blocks the cytotoxic activity of the isolate. While established cell lines are probably the most efficient source of material for evaluating the cytotoxic activity of clinical isolates, primary cells derived from patient biopsies may have value in determining the effectiveness of various antibiotic treatments on such activity. For example, chronic wounds are often infected with multiple species of bacteria and can, in some cases, persist for months. A primary cell line established from a biopsy taken at the wound site can potentially be used to determine which antibiotics block the invasive and cytotoxic phenotypes of isolates derived from the wound. This may enable the physician to use antibiotics to block bacterial spread and tissue damage and may potentially expand the use of the world’s rapidly diminishing antibiotic formulary.
Conclusion
Phenotypic data beyond basic biochemical activity is currently not captured in the clinical microbiology laboratory. However, the phenotypic evaluation of clinical isolates can provide the clinician with an expanded data set and provide insights into the biology of the offending organism that can improve clinical decision making. The ability to determine whether or not an organism is cytotoxic will better enable the clinician to decide whether or not to pursue aggressive antibiotic therapy and whether or not to pursue aggressive surgical debridement. The ability to monitor bacterial-mediated cytotoxicity in real time will enable antibiotics to be evaluated for their ability to block the cytotoxic and invasive phenotypes of bacterial pathogens and has the potential to expand the use of the current antibiotic inventory.
While bacterial cytotoxicity has been evaluated in the research setting, it is not currently evaluated in the clinical microbiology laboratory. ECIS was developed as a novel means of monitoring the behavior of mammalian cell monolayers by recording impedance values as a function of time. ECIS technology provides an automated rapid means of evaluating cell behavior. The ability of ECIS to monitor the cytotoxic effects of viral pathogens on mammalian cells has been previously demonstrated, and the ability of this technology to monitor bacterial pathogenesis is currently being evaluated. Although impedance methods are not typically applied in the clinical microbiology laboratory at this time, biological impedance analysis and ECIS are mature technologies that can be readily applied to the hospital setting.
The views expressed in this article are those of the authors and do not reflect the official policy or position of the Department of the Army, Department of Defense, or the U.S. Government.
REFERENCES
- Reller LB, Weinstein MP, Peterson LR et al.. Role of clinical microbiology laboratories in the management and control of infectious diseases and the delivery of health care. Clin Infect Dis. 2001;;32(4):605-610.
- Association of Public Health Laboratories. 2015. Specimen processing in clinical microbiology. What’s new? [PowerPoint slides]. http://eo2.commpartners.com/users/APHL/downloads/588-905-15-6SlidesPerPage.pdf.
- Köser CU, Ellington MJ, Cartwright EJ, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS pathogens. 2012; 2;8(8):e1002824.
- Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest J. 2000;118(1):146-155.
- Kollef MH, Ward S, Sherman G, et al.Inadequate treatment of nosocomial infections is associated with certain empiric antibiotic choices. Crit Care Med. 2000;28(10):3456-3464.
- Campbell CE, Laane MM, Haugarvoll E, Giaever I. Monitoring viral-induced cell death using electric cell–substrate impedance sensing. Biosensors and Bioelectronics. 2007;23(4):536-542.
- McCoy MH, Wang E. Use of electric cell-substrate impedance sensing as a tool for quantifying cytopathic effect in influenza A virus infected MDCK cells in real-time. J Virolog Methods. 2005;130(1):157-161.
- El Asmar R, Panigrahi P, Bamford P et al.. Host-dependent zonulin secretion causes the impairment of the small intestine barrier function after bacterial exposure. Gastroenterology. 2002;123(5):1607-1615.
- Spitz JA, Yuhan RO, Koutsouris AT, Blatt CA, Alverdy JO, Hecht GA. Enteropathogenic Escherichia coli adherence to intestinal epithelial monolayers diminishes barrier function. Amer J Physiology-Gastrointestinal and Liver Physiology. 1995;268(2):G374-379.
- Bayles KW, Wesson CA, Liou LE, Fox LK, Bohach GA, Trumble WR. Intracellular staphylococcus aureusescapes the endosome and induces apoptosis in epithelial cells. Infection and Immunity. 1998;66(1):336-342.
- Buommino E, Morelli F, Metafora S, et al. Porin from Pseudomonas aeruginosainduces apoptosis in an epithelial cell line derived from rat seminal vesicles. Infection and Immunity. 1999 Sep 1;67(9):4794-4800.
- Paesold G, Guiney DG, Eckmann L, Kagnoff MF. Genes in the Salmonella pathogenicity island 2 and the Salmonella virulence plasmid are essential for Salmonella‐induced apoptosis in intestinal epithelial cells. Cellular microbiology. 2002;4(11):771-781.
- Soong G, Parker D, Magargee M, Prince AS. The type III toxins of Pseudomonas aeruginosa disrupt epithelial barrier function. J Bacteriology. 2008;190(8):2814-21.
- Kannan TR, Baseman JB. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. PNAS. 2006;103(17):6724-6729.
- Kasendra M, Barrile R, Leuzzi R, Soriani M. Clostridium difficile toxins facilitate bacterial colonization by modulating the fence and gate function of colonic epithelium. J Infect Dis. 2013;209(7):1095-1104.
- Ohnemus U, Kohrmeyer K, Houdek P, et al. Regulation of epidermal tight-junctions (TJ) during infection with exfoliative toxin-negative Staphylococcus strains. J Invest Dermatol. 2008;128(4):906-916.
- Baron C, Coombes B. Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infectious Disorders-Drug Targets (formerly Current Drug Targets-Infectious Disorders). 2007 Mar 1;7(1):19-27.
- Tran SL, Puhar A, Ngo-Camus M, Ramarao N. Trypan blue dye enters viable cells incubated with the pore-forming toxin HlyII of Bacillus cereus. PLoS One. 2011;6(9):e22876.
- Palma PF, Baggio GL, Spada C, Silva RD, Ferreira SI, Treitinger A. Evaluation of annexin V and Calcein-AM as markers of mononuclear cell apoptosis during human immunodeficiency virus infection. Brazilian Journal of Infectious Diseases. 2008; 12(2):108-114.
- Vega-Avila E, Pugsley MK. An overview of colorimetric assay methods used to assess survival or proliferation of mammalian cells. InProc West Pharmacol Soc. 2011; 54(10): e4).
- Xiao C, Luong JH. On‐line monitoring of cell growth and cytotoxicity using electric cell‐substrate impedance sensing (ECIS). Biotechnology Progress. 2003;19(3):1000-1005.
- Wegener J, Keese CR, Giaever I. Electric cell–substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exper Cell Res. 2000;259(1):158-166.
- Pfaller MA, Herwaldt LA. The clinical microbiology laboratory and infection control: emerging pathogens, antimicrobial resistance, and new technology. Clinical Infect Dis. 1997;25(4):858-870.
- Franklin C, Liolios L, Peleg AY. Phenotypic detection of carbapenem-susceptible metallo-β-lactamase-producing gram-negative bacilli in the clinical laboratory. J Clin Microbiol. 2006;44(9):3139-3144.
MAJ Michael A. Washington, PhD, M(ASCP) and Carmen E. Campbell, MSc, serve in the Department of Clinical Investigation, Tripler Army Medical Center, Honolulu, HI.