Applications of nucleic acid testing in diagnosis and therapy
Nucleic acid testing or nucleic acid amplification testing, often abbreviated as NAT or NAAT, is a technique that involves amplification and detection of genetic material—the nucleic acids, DNA or RNA—for diagnosis or to provide guidance on therapy. Though the genetic material of every living being is composed of DNA or RNA, variations exist in genome sequences. This genetic variation makes NAT an ideal technique for identification of infectious diseases, cancer, genetic disorders, and mitochondrial disorders and as an aid to personalized and precision medicine based on the knowledge of pharmacogenomics.
NAT can be performed on various types of clinical specimens depending on the disease being diagnosed. Blood, plasma, serum, cerebrospinal fluid, sterile body fluids, sputum, urine, stool, and tissues are some specimens commonly used for NAT. The nucleic acids are first extracted from the cell and other cellular components either by manual or automated methods. The extracted nucleic acids can then be amplified and detected or sequences read using different methods—polymerase chain reaction (PCR), real-time PCR, microarrays, sequencing (Sanger and next-generation), etc. With the advancement in the technology for amplification and detection or sequencing, there has been a great transformation in the applications of NAT. The major applications are discussed below.
NAT for blood screening
NAT has been associated with blood screening for some time. It was first introduced by the German Red Cross in 1997 for blood screening to reduce the risk of transfusion-transmitted viral infections due to the failure of serologic screening tests to detect recently infected donors in the pre-seroconversion “window” phase of infection.1 With serology tests, it takes about two months after infection for hepatitis C virus (HCV) antibodies to be detected, while NAT can detect HCV RNA about five days after infection. To reduce the window period for detection, European Union regulators began to require in 1999 that all plasma be tested by NAT techniques for HCV if derivatives made from such plasma were to be sold in Europe.
This announcement was a major impetus for the development and implementation of NAT of blood and plasma from donors in the United States and other developed countries. In the U.S., NAT was initially used to screen HCV and human immunodeficiency viruses types 1 and 2 (HIV 1, 2) in blood and blood products, and subsequently extended to hepatitis B virus (HBV) and West Nile virus (WNV). Screening for human T-lymph tropic virus types I and II (HTLV-I, II) and Treponema pallidum (causing syphilis) are also routinely performed using the serological method only. NAT has been adopted in several other countries around the world, including Canada, France, Australia, New Zealand, South Africa, and other countries in Europe and Asia.2,3 In addition to screening for the organisms mentioned above, blood screening for other organisms, including Plasmodium vivax and Plasmodium falciparum (causing malaria), and Trypanosoma cruzi (causing Chagas disease) are also recommended, depending on the local epidemiological evidence of the disease.4
NAT in detecting infectious diseases
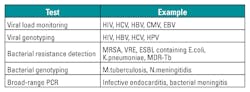
NAT is extensively used to detect and identify organisms for proper diagnosis, prognosis, and treatment of diseases. Diagnosing infectious diseases by detecting the nucleic acid of the causative agent in clinical specimens has been found to be more rapid, sensitive, and specific compared to the traditional method of diagnosis using culture or immunological methods. Table 1 provides some examples of organisms that have been detected by NAT in clinical laboratories.5
In virology, in addition to detecting the viruses, NAT further aids in identifying genotypes and subtypes, determining resistance to particular antibiotics, and measuring viral load. These applications help to provide guidance on treatment. In bacteriology, NAT has also been applied to resistance testing, the detection of infection due to fastidious bacteria, and the detection of bacterial infection after antibiotics have been administered.
NAT also helps in epidemiological studies and infection control. Table 2 provides some examples of applications of NAT other than detection.5
NAT in predicting cancer and guiding cancer treatment
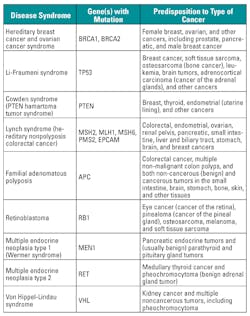
Cancer is a disease caused by an uncontrolled division of abnormal cells in a part of the body. With improved sequencing capability and knowledge of the human genome (DNA), traditional descriptions of cancer—for example, by the organ of primary occurrence—are being superseded by a new classification framework that focuses on the genetic abnormalities and molecular derangements of malignant tumors. The genetic-level changes may consist of single or multiple sequence changes in cell DNA (mutations), addition or deletion of DNA, or changes in the number of copies of particular DNA sequences in cancer cells. Nucleic acid testing helps to identify genetic variations and predicts predisposition to cancer, alters diagnostic categories, enhances treatment strategies, enables early detection and prevention, and improves outcomes for cancer patients. Nucleic acid testing has led to the emergence of precision and personalized medicine—that is, the tailoring of treatment based on the individual’s genetic make-up.
Table 3 provides a partial list of predisposition to certain type of cancer based on gene mutations. NAT helps in detection of the mutations in the genes.6
Table 4 provides a partial list of cancer drugs that are indicated for cancer patients with particular genetic variants. NAT helps in detection of the patient’s genetic mutations.7
NAT for personalized and precision medicine
With today’s increased knowledge of pharmacogenomics, nucleic acid testing is being increasingly used to provide personalized and precision medicine and avoid adverse reactions from drugs in individuals.
To define the terms: pharmacogenetics studies the effect of a single gene on drug response, while pharmacogenomics deals with the effects of multiple genes on drug response. The term pharmacogenetics was coined by Vogel in 1959,8 but most of the progress in pharmacogenetics has been made in recent years. Researchers and clinicians have come to understand that every individual metabolizes drugs according to his or her genetic variants and therefore responds to drugs variably. This has enabled advancements in personalized and precision medicine.
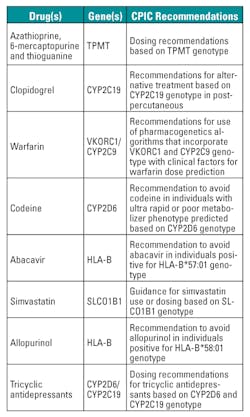
Over the last decade, the U.S. Food and Drug Administration (FDA) has been aggressive in providing genetic labeling on new drugs, and also updating product labels for a number of existing therapies, such as warfarin and 6-mercaptopurine. At present, some 120 drugs9 have pharmacogenetics information in their FDA product label. A current listing of all drugs with such information in the product label can be found in U.S. FDA Table of Pharmacogenomics Biomarkers in Drug Labels.10 Pharmacogenetics information in product labels ranges from boxed warnings, the highest level of warning in the product label, to information in the clinical pharmacology section.
The Clinical Pharmacogenetics Implementation Consortium (CPIC), formed in 2009, sets guidelines on drug therapy based on pharmacogenetics information.11 By early 2013, eight CPIC guidelines had been published, which include TPMT and thiopurines,12 CYP2C19 and clopidogrel,13 VKORC1/CYP2C9 and warfarin,14 CYP2D6 and codeine,15 HLA-B and abacavir,16 SLCO1B1 and simvastatin,17 HLA-B and allopurinol,18 and CYP2D6/CYP2C19 and tricyclic antidepressants.19 These guidelines are summarized in Table 5 . Production of a number of other guidelines is ongoing and a listing of in-progress guidelines can be found at The Pharmacogenomics Knowledgebase.20
NAT in screening for genetic disorders
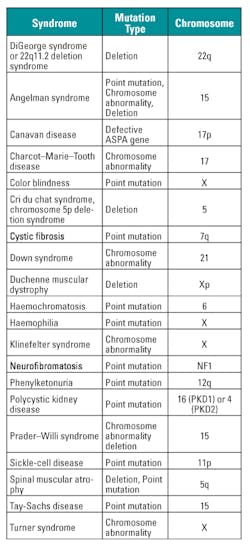
NAT is being extensively used for prenatal screening and carrier screening for genetic disorders. Congenital anomalies account for 276,000 perinatal deaths by pregnancy Week 4 annually on a global basis.21 The aim of prenatal screening is to detect birth defects, such as neural tube defects; chromosome abnormalities (e.g., Down syndrome, fragile X syndrome); and genetic disorders and other conditions (e.g., spina bifida, cleft palate, Tay Sachs disease, sickle cell anemia, thalassemia, cystic fibrosis, and muscular dystrophy).21
Carrier screening, the testing of parents in preparation for pregnancy, is used to identify genetic mutations that could cause serious inherited disorders. Some of the more common disorders for which such screening is done are cystic fibrosis, sickle cell disease, thalassemia, and Tay-Sachs disease.21
In the Ashkenazi (Eastern European descent) Jewish population, a number of genetic disorders occur more frequently than in the general population. It has been estimated that one in four individuals is a carrier of one of several genetic conditions. These diseases include Tay-Sachs disease, Canavan, Niemann-Pick, Gaucher, familial dysautonomia, Bloom syndrome, Fanconi anemia, cystic fibrosis, and mucolipidosis IV. Table 6 provides a list of common genetic disorders.
NAT in diagnosis of mitochondrial diseases
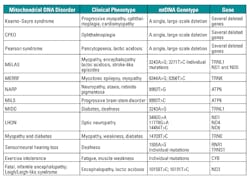
Mitochondrial diseases are a group of disorders caused by dysfunctional mitochondria—the cellular organelle in which respiration and energy formation occur. Mitochondrial diseases can be caused by genetic mutation in the mitochondrial DNA (mtDNA) or in the nuclear DNA. Mitochondrial diseases may affect a single organ—for example, the eye in Leber hereditary optic neuropathy (LHON)—or may involve many organs, and they often result in major neurologic and myopathic symptoms. Table 7 lists a few disorders caused due to mutations in mitochondrial DNA.22
The genetic basis of mitochondrial disease was first discovered in the late 1980s. Since then, mitochondrial disease diagnosis has continued to evolve with improvement in NAT technology. Now, with NAT technologies such as sequencing and deletion analysis of an increasing number of individual nuclear genes linked to mitochondrial disease, genome-wide microarray analysis for chromosomal copy number abnormalities and mitochondrial DNA whole genome sequence analysis have greatly helped in understanding and diagnosing mitochondrial diseases.23
Conclusion
Nucleic acid testing is growing very rapidly in laboratory medicine. With the emergence of newer technology and easy-to-use sample-to-result systems, NAT can be applied in more areas of testing. In the future, NAT will play an important role in making personalized and predictive medicine the standard of care for better healthcare.
REFERENCES
- Hourfar MK, Jork C, Schottstedt V, et al. Experience of German Red Cross blood donor services with nucleic acid testing: Results of screening more than 30 million blood donations for human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus. Transfusion. 2008;48:1558–1566.
- Busch MP, Glynn SA, Stramer SL, et al. A new safety strategy for estimating risks of transfusion-transmitted viral infections based on rates of detection of recently infected donors. Transfusion. 2005;45:254-264.
- Chamberland ME, Alter HJ, Busch MP, Nemo G, Ricketts M. Emerging infectious disease issues in blood safety. Emerging Infectious Diseases. Vol. 7, No. 3 Supplement, June 2001.
- World Health Organization. Screening donated blood for transfusion-transmissible infections. http://www.who.int/bloodsafety/ScreeningDonatedBloodforTransfusion.pdf.
- Speers DJ. Clinical applications of molecular biology for infectious diseases. Clin Biochem Rev. 2006 27(1): 39–51.
- National Cancer Institute. Genetic testing for hereditary cancer syndromes. http://www.cancer.gov/about-cancer/causes-prevention/genetics/genetic-testing-fact-sheet.
- Personalized Medicine Coalition. The case for personalized medicine. 4th edition. 2014. http://www.personalizedmedicinecoalition.org/Userfiles/PMC-Corporate/file/pmc_the_case_for_personalized_medicine.pdf.
- Vogel F. [Modern problems of human genetics.] Ergeb Inn Med Kinderheilkd. 1959;12:52-125. In German.
- Singer DE, Albers GW, Dalen JE et al. Antithrombotic therapy in atrial fibrillation: the Seventh ACCP Conference on Antithrombotic and Thrombolytic Therapy. Chest. 2004;126(3, suppl):429S-456S.
- US FDA Table of Pharmacogenomic Biomarkers in Drug Labels. www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm.
- Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464-467.
- Relling MV, Gardner EE, Sandborn WJ, et al., Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89(3):387-391.
- Scott SA, Sangkuhl K, Gardner EE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90(2):328-332.
- Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90(4):625-629.
- Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91(2):321-326.
- Martin MA, Klein TE, Dong BJ, Pirmohamed M, Haas DW, Kroetz DL, Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin Pharmacol Ther. 2012;91(4):734-738.
- Wilke RA, Ramsey LB, Johnson SG, et al. The Clinical Pharmacogenomics Implementation Consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92(1):112-117.
- Hershfield MS, Callaghan JT, Tassaneeyakul W, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clin Pharmacol Ther. 2013;93(2):153-158.
- Hicks JK, Swen JJ, Thorn CF, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013;93(5):402-408.
- The Pharmacogenomics Knowledgebase. www.PharmGKB.org.
- Gabrielle Gabrielle Heilek. Nucleic Acids: The Use of Nucleic Acid Testing in Molecular Diagnostics Chapter 5. http://www.intechopen.com/books/howtoreference/nucleic-acids-from-basic-aspects-to-laboratory-tools/nucleic-acids-the-use-of-nucleic-acid-testing-in-molecular-diagnostics.
- Robert W. Taylor and Doug M. Turnbull Mitochondrial DNA Mutations In Human Disease Nat Rev Genet. 2005 May;6(5):389–402.
- McCormick E1, Place E, Falk MJ. Molecular genetic testing for mitochondrial disease: from one generation to the next. Neurotherapeutics. 2013 Apr;10(2):251-61.
Rajasri Chandra, MS, MBA, serves as Product Manager for molecular diagnostics provider AutoGenomics, Inc., based in Vista, CA.
About the Author

Rajasri Chandra, MS, MBA
is a global marketing leader with expertise in managing upstream, downstream, strategic, tactical, traditional, and digital marketing in biotech, in vitro diagnostics, life sciences, and pharmaceutical industries. Raj is an orchestrator of go-to-market strategies driving complete product life cycle from ideation to commercialization.