The promise of personalized healthcare—matching the right therapy to the right patient—rests with high-value diagnostics that can guide “which patients” will respond to “which therapy” within a specific disease. Recent clinical successes in the area, however, have generally been limited to diseases that have specific mutations in a single gene.
It is now widely accepted that most chronic diseases, including asthma, diabetes, Parkinson’s disease, late-onset Alzheimer’s disease, and most cancers, are far too complex to rely on genomic variations alone. Instead, the molecular drivers of disease are in fact manifested across thousands of interrelated, biochemical pathways, and these disease drivers need to be thoroughly interrogated in order to provide effective treatment options.
Recent advances in our understanding of human genomics had been expected to, in turn, increase our understanding of the etiology and pathogenesis of common diseases. In time, these advances were also expected to deliver opportunities for the prevention, early detection, and treatment of disease, in part by enabling healthcare providers to implement individualized preventive and therapeutic strategies based on patients’ genomic profiles. However, despite the completion of the Human Genome Project and the success of recent genome-wide association studies (GWAS), the usefulness of genomic information alone in treating or predicting disease has been limited and applicable only to a small fraction of patient population in certain diseases. Furthermore, because most common diseases stem from complex interactions among multiple genetic and non-genetic factors, each of which confers only minor increases in risk, the predictive value of genome-limited testing or profiling may be inadequate as a useful platform for therapeutic intervention.
The widely touted field of molecular diagnostics (MDx) is concerned with multiple platforms and tests that rely on the detection and/or analysis of discreet packets of genetic material (RNA or DNA-based). MDx tests started to be adopted clinically in 2000 and were initially focused on infectious diseases. Since then, early insights into the mechanisms of human disease, provided in part by gene expression profiling experiments, have led to a number of MDx tests being developed for cancer. However, with the increasing realization that disease biology is highly complex and extends well beyond genetic information, the future widespread use of MDx tests in personalized medicine is uncertain.
Systems diagnostics—a new generation of diagnostics
In order to deliver true personalized medicine and solve the problem of disease complexity, physicians will require diagnostics that holistically integrate and interpret multiple types of biological data. Systems diagnostics (SysDx), in contrast to MDx, are diagnostic tests that incorporate a wide series of biomarkers from different biological disciplines. Derived from large, multi-omic databases, SysDx tests consist of biomarkers that are selected through a rigorous analysis of a patient’s biological map. SysDx can include genomic, epigenomic, transcriptomic, proteomic, metabolomic, and electronic medical record information. This comprehensive biomarker analysis is then used to generate a clinically relevant diagnostic report that physicians and patients can use to make an optimal treatment decision. Due to its holistic “multi-omic” approach, SysDx has significant applicability in multi-factorial diseases.
As described, a major differentiating factor in the development of SysDx tests compared to more traditional approaches is the capability to incorporate and analyze ALL relevant molecular information in a disease, across thousands of patients, to identify biomarkers that are linked to disease-driving mechanisms. The true genesis of SysDx tests stems back to years of collaborative biomarker discovery work between specialized systems biology firms and the pharma & biotech industry. There are many machine learning methods that have been applied to biomarker discovery; however, the best approaches rely on matching known patterns of multiple biomarkers, as these avoid problems associated with pattern over-fitting. Importantly, because SysDx is based on well-annotated, prior scientific knowledge, the approach can go beyond identifying new relationships among biomolecular entities.
Having access to biological knowledge databases is a critical substrate for developing “Big Data” solutions for personalized healthcare applications. Ideally, these platforms are scalable and extensible so that the development of new tests can address many opportunities. Databases can be sourced either privately or through open, public consortiums such as the OpenBEL community (www.openbel.org), which is supported by the Linux Foundation.
The development of SysDx tests starts with the contribution of patient data from a number of sources including academic institutions, pharmaceutical and biotech companies, and commercial content providers.
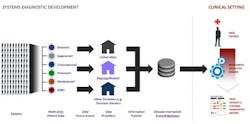
With the ability to identify biomarkers before the patient data is applied, SysDx tests can then be independently developed according to several discrete steps (Figure 1):
- Identify the major mechanisms active in a specific disease
- Develop biomarkers that identify patients with each disease mechanism
- Understand how mechanisms differ within a disease
- Refine/employ biomarkers to create predictive diagnostic tests
Figure 1. Systems diagnostics development process and application in clinical settings.
SysDx can be used as clinically relevant diagnostic reports that can inform treatment decisions for a patient, or for pharmaceutical companies to use as a companion diagnostic to stratify patient populations. Compared to current state-of-the-art MDx testing, SysDx testing provides numerous benefits to patients, physicians and also payers, by addressing unfavorable healthcare economics. The SysDx approach, for example, is disease agnostic and is easily scalable to address much larger groups of patients. In addition, SysDx can comprehensively break down a single complex disease into multiple, “mechanism-addressable” targets—that is, by taking one cancer type and “orphanizing” the disease into multiple subcategories with tailored treatment options. On the health economics side, SysDx are positioned to be very attractive to payers as they are able to optimize patient treatment paradigms. SysDx tests will remove the high costs associated with patients being prescribed ineffective and potentially harmful drugs and identify the correct therapy that will best return the patient to a healthy state.
Most significantly, these tests have the potential to dramatically improve the quality of life for millions of patients who currently suffer due to lack of information that can be provided by SysDx tests.
SysDx for rheumatoid arthritis
In the case of rheumatoid arthritis (RA), it is paramount that accurate early-stage diagnosis of RA patients be conducted. If left untreated, it is expected that 70% of patients will go on to develop irreversible joint damage within two years of presenting with symptoms.
Over the last 10 years, significant progress has been made in the treatment of RA; however, many patients remain partial responders to the marketed therapies, and true remission from disease symptoms is rarely achieved. Therefore, there is a major need for personalized tests that will predict whether patients will or will not respond prior to an arthritis therapy. Today, only approximately 40% of RA patients clinically benefit from the standard of care biologics therapies, and identifying non-responders early would limit disease progression.
Physicians typically use a combination of clinical assessment, x-rays, and laboratory tests to help determine the risk of progression of structural damage in RA patients. Serologic factors, such as rheumatoid factor (RF), anti-CCP antibodies, and an elevated baseline erythrocyte sedimentation rate (ESR) or C-reactive protein (CRP) level, are linked with disease progression; however, none of these tests actually provides disease- or patient-specific information. In addition, as is typical of many autoimmune diseases, there are many different types of biological factors (e.g., genes, enzymes, metabolites, etc.) involved, and focusing on a single class alone won’t provide adequate patient diagnosis. Given the multi-omics analysis capabilities of the SysDx platform, it is very suitable and well positioned to tackle these complex diseases.
With the advent of big data in healthcare, there is a significant opportunity to leverage increasingly commoditized platform technologies such as gene expression arrays, next generation sequencing, platform proteomics, and electronic medical records. All the information from these sources can be assimilated by SysDx discovery platforms. SysDx reporting will be evidence-based and will provide probabilistic information that will aid physicians and patients in making important decisions about appropriate therapy.
SysDx’s ability to harness multiple types of molecular information to derive clinically relevant multi-omic biomarkers will unlock the potential of “big data” as applied to personalized patient care.