Trends for early detection of aneuploidy and trisomy
Early detection of aneuploidy (an abnormal number of chromosomes) during the first trimester of pregnancy provides essential information to expectant families and their healthcare providers. Aneuploidy is the most frequent genetic disorder observed in live births and miscarriages; trisomies are the most prevalent, accounting for approximately 53% of all chromosome abnormalities. Advances in healthcare and educational approaches have greatly improved the quality of life and life expectancy of individuals born with trisomy (three copies of a given chromosome instead of a pair). The most common forms of autosomal trisomy are trisomy 21, which results in Down syndrome, and trisomy 18, which results in Edwards Syndrome. In rare cases, a fetus with trisomy 13 (Patau syndrome) can survive. Autosomal trisomy is frequently associated with severe congenital abnormalities, mental retardation, and shortened life expectancy. Aneuploidy of sex chromosomes can also occur: The presence of extra X chromosome(s) causes Klinefelter syndrome in men and Triple X syndrome in women, while monosomy X gives rise to women with Turner syndrome.
The techniques for aneuploidy detection have evolved from cytological approaches prior to the 1990s, to molecular DNA tests (Qualitative Fluorescence Polymerase Chain Reaction, or QF-PCR) in the mid-1990s, to exciting, new techniques utilizing next generation sequencing that are currently under development for improvement of early aneuploidy detection.
Amniocentesis is the most common method to obtain a prenatal fetal sample for trisomy and aneuploidy testing. Amniocentesis is not recommended for all pregnancies, as it carries a risk of causing miscarriage. The most common guidelines for when to perform this prenatal testing include the following: an abnormal alpha-fetoprotein (AFP) test; the expectant mother being over the age of 35 (the age at which the risk of miscarriage from the amniocentesis is approximately equal to the probability that the fetus has an abnormal chromosome number); or a family history of a birth defect for which a diagnostic test is available.1 Improved, accurate, non-invasive aneuploidy tests are anticipated in the future, as an estimated 75% of all trisomic births are to women under the age of 35.
Traditionally, aneuploidy of fetal cells was detected from a karyotype of cultured cells or by fluorescent in situ hybridization (FISH) of uncultured cells. A karyotype compares banding patterns of stained chromosomes from actively dividing cells. FISH studies detect commonly occurring aneuploidies with multicolor, commercially available, chromosome specific probes for chromosomes 13, 18, 21, X, and Y.2 Cytological tests can detect an abnormal chromosome number, but they are time-consuming, require counting of chromosomes in several cells, may require two independent analysts to interpret the results, and are not suitable for automation or accurate high-throughput analysis.
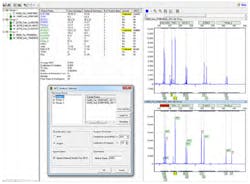
QF-PCR is more commonly used to detect aneuploidy in prenatal samples.3,4 This technique is quantitative; additionally, the laboratory and data analysis steps can be partially automated for accurate results of more samples in less time. QF-PCR uses chromosome specific primers to amplify DNA fragments for each of the chromosomes of interest. DNA fragments are separated with capillary electrophoresis and analyzed by fragment size and number of fragments. The peak height, or relative fluorescence, is used to quantify the amount of DNA amplified, determining if the expected pair of each chromosome or aneuploidy is present. Three DNA fragments, visualized as peaks of approximately the same height in an electropherogram, or peak ratios of 2:1 and 1:2, are indications that the sample is in the trisomic range.
Custom chemistries or commercial PCR kits provide primer sets that will detect chromosomes 13,18, 21, X, and Y. In many cases these multiplexes are sufficient for conclusively detecting and quantifying aneuploidy.
The peak pattern shown in Figure 1 is an example of a sample that is in the trisomic range for Down syndrome. The sample in Figure 2 has one peak ratio that is in the inconclusive range for chromosome 21. Values falling in the ranges (1.4-1.8 and 0.65-0.8) lying outside the normal and triallelic ranges are referred to as intermediate ranges and the ratio calculations are referred to as inconclusive results. Such results can usually be resolved by re-testing the sample.5 Most will test samples that fall in the inconclusive range with a separate multiplex containing additional primers for that chromosome. Best practice guidelines also recommend that all samples undergoing aneuploidy testing are first evaluated for maternal cell contamination (MCC).6 Amniotic samples with MCC can be misinterpreted as trisomic (Figure 3). QF-PCR increases the efficiency of aneuploidy testing but is limited by the number of primer sets that amplify segments of the chromosomes of interest.
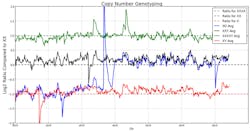
Next-generation (NGS) DNA sequence analysis of cell-free fetal DNA (cffDNA) obtained from maternal blood is a promising upcoming technology.7 Many amplicons across the chromosome are sequenced; providing more detailed results from deep sequencing. The graph in Figure 4 illustrates results from four samples. Three samples are within the normal range for chromosome 21; one sample is within the trisomic range. Figure 5 is a summary of the results of four samples screened for X chromosome aneuploidy. Ranges for X, XX, and XXXX are indicated by a dashed line. The results are indicated by solid lines and indicate a normal XY male, a male with an additional X chromosome (XXY), a male with three additional X chromosomes (XXXXY) and a female with a missing X chromosome (X0). Barcoding will enable processing hundreds of samples, increasing efficiency and decreasing cost of aneuploidy screening with the high-throughput capabilities of NGS.
References
- Encyclopedia of Surgery: Amniocentesis. http://www.surgeryencyclopedia.com/A-Ce/Amniocentesis.html. Accessed November 28, 2012.
- Toth T, Findlay I, Papp C, et al. Prenatal detection of trisomy 21 and 18 from amniotic fluid by quantitative fluorescent polymerase chain reaction. J Med Genet. 1998;35:126-129.
- Stumm M, Wegner RD, Bloechle M, Eckel H. Interphase M-FISH applications using commercial probes in prenatal and PGD diagnostics. Cytogenetic and Genome Research. 2006;114:296-301.
- Brown L, Abigania M, Warburton D, Brown S. Validation of QF-PCR for prenatal aneuploidy screening in the United States. Prenat Diagn. 2006;26:1068-1074.
- Association for Clinical Cytogenetics and Clinical Molecular Genetics Society QF-PCR for the Diagnosis of Aneuploidy Best Practice Guidelines. 2007 and 2012.
- Allen S, Moutford R, Butler A, Mann K, Treacy B. Practice guidelines for the testing for maternal cell contamination (MCC) in prenatal samples for molecular studies. Clinical Molecular Genetics Society. 2008.
- Nepomnyashchaya YN, Artemov AV, Roumlantsev SA, Roumyantsev AG, Zhavoronkov A. Non-invasive prenatal diagnostics of aneuploidy using next-generation DNA sequencing technologies, and clinical considerations. Clin Chem Lab Med. 2012;Sept 29:1-14.
Teresa Snyder-Leiby, PhD, is Product Manager for SoftGenetics LLC, manufacturer of genetic analysis software GeneMarker®, NextGENe®, Mutation Surveyor®, and ChimerMarker™.