To earn CEUs, see current test at
www.mlo-online.com
under the CE Tests tab. The September test covers all articles in this
section, except the product announcement.
LEARNING OBJECTIVES
Upon completion of this article, the
reader will be able to:
- Identify organisms that commonly cause HAIs and ARIs, including
Clostridium difficile-associated infection (CDI). - Name mechanisms for the increases in HAIs, ARIs, and CDI.
- Name trends in HAIs, ARIs, and CDI.
- Name types of infections that put patients at greater risk of
fatality from HAIs and ARIs. - Describe procedures to reduce the number of HAIs, ARIs, and CDI.
- Understand testing methods for HAIs, ARIs, and CDI.
Reducing HAIs and ARIs: partnering with clinical labs
Many of the most
insidious of healthcare-associated (or hospital-acquired) infections
(HAIs) — and antibiotic-resistant infections (ARIs) — are creatures of
our own unintelligent design. Methicillin-resistant Staphylococcus
aureus (MRSA), vancomycin-resistant enterococci (VRE), and a growing
number of other pathogens developing resistance to many antibiotics are
directly attributable to the overuse and misuse of these drugs. For
example, 75% of antibiotics are prescribed for acute respiratory-tract
infections, despite the fact that approximately 80% of them are of viral
origin.
In just over a decade, S aureus, once
described as a “controllable nuisance,” has evolved into MRSA, one of
the fastest-growing resistant infections that does not respond to most
antibiotics. In the United States, current MRSA rates exceed 50%1
of all S aureus infections and stand at nearly 90% in some Asian
countries.2 Lack of compliance with hand-disinfection
procedures, inappropriate use of antimicrobials, and underlying diseases
prior to hospitalization are some of the most common ways MRSA is
spread. In the past five years, MRSA has exploded in the general
community — an alarming and ominous trend.
In 1993, there were fewer than 2,000 MRSA
infections in U.S. hospitals. By 2005, the figure had shot up to
368,000, according to the Agency for Healthcare Research and Quality. At
Morton Plant Hospital, we now see HAIs almost every day. It is estimated
that about 70% of bacteria that cause infections in hospitals are
resistant to at least one of the drugs most commonly used to treat
infections.
Those who track the genetic shift and drift that
makes these pathogens so adaptable believe that VRE poses the next
serious health threat. Unfortunately, these scientists believe that the
organism has transferred a key antibiotic-resistance gene to
Staphylococcus. We are also seeing more cases of Klebsiella
pneumoniae Carbapenemase- (KPC-) producing organisms, such as
Escherichia coli and Salmonella. KPC pathogens are virtually
impervious to all penicillins, cephalosporins, carbapenems, and
axteonam, which leaves us with no available treatment.
MRSA plus H1N1 influenza A a threat
At the Second World HAI Forum held in last month
in the Les Pensi`eres Conference Center in Veyrier-du-Lac, France, these
supercharged microorganisms were discussed by experts from around the
globe, gathered to anticipate what their next move will be. These
experts focused on two looming threats.
In just over a decade, S aureus, once described as a
“controllable nuisance,” has evolved into MRSA, one of the
fastest-growing resistant infections that does not respond to most
antibiotics.
The first is the convergence of MRSA and H1N1
influenza A. When combined with MRSA, even mild seasonal flu can become
very dangerous. The virus distracts the immune system, which has a more
difficult time battling the bacterial infection that can lead to severe
pneumonia. A 50% mortality rate has been reported with
community-acquired MRSA pneumonia.2 This has already been
seen in Australia, which is coming to the end of its flu season.
Fortunately, these cases were not common; however, when they did occur,
they were frequently fatal. The extent of the problem in the Northern
Hemisphere will be determined by the severity of the H1N1 pandemic and
the efficacy of the vaccine.
The second looming threat identified at the
recent World HAI Forum are bacteria that produce extended spectrum
beta-lactamase, or ESBL. This enzyme has evolved the ability to render
many antibiotics useless. ESBLs are produced by E coli and K
pneumoniae, which are becoming more pervasive and difficult to treat
in the hospital setting. In fact, K pneumoniae Carbapenemase can
inactivate nearly all antibiotics, including carbapenems, which had been
the medical “weapon of last resort.”
Resistance enzymes that bypass extended spectrum
cephalosporin and carbapenem antibiotics are known as carbapenemases.
These molecules have versatile hydrolytic
capacities that inactivate antibiotics in the penicillin, cephalosporin,
monobactam, and carbapenem families.
Still, doctors perpetuate the problem by
increasing the prescription of carbapenems due to the spread of
pathogens armed with these resistance enzymes, thereby inadvertently
creating carbapenemase-producing bacteria resistant to the antibiotic.
One of the central battlegrounds in the efforts
to overcome antibiotic resistance is the human lung, which is the
primary point of entry for many of these pathogens. Each year,
235 million doses of antibiotics are prescribed, but
between 20% to 50% of these prescriptions are unnecessary.3,4
Of the 41 million antibiotic prescriptions written in the United States
each year for respiratory infections, as many 22.5 million (55%) are
likely to have been prescribed for non-bacterial infections.5
One way to dramatically reduce overuse of
antibiotics is to avoid treating viral infections and simple
inflammation, as in the cases of asthma and chronic obstructive
pulmonary diseases (COPD) with antibiotics that do no good.
C difficile is a Gram-positive anaerobic bacillus that exists in
vegetative and spore forms, and is spread through the fecal-oral route.
Two recent studies demonstrate both the impact of
this promiscuous use of antibiotics and the benefits that can be
realized if we “kick this habit.” Researchers at the Cook County
Hospital in Chicago published research this month on the true cost of
antibiotic-resistant infections.6
They concluded that the healthcare costs associated with ARIs in that
hospital in 2000 ranged between $18,000 to $29,000 per patient, and
these patients remained hospitalized for an additional 6.4 to 12.7 days
in order to have these infections treated. These patients were more than
twice as likely to die than comparable patients who did not become
infected with antibiotic-resistant organisms. This study was one of the
first to also look at the societal costs of ARIs — those costs borne by
the patients and their families — resulting from lost wages or, in the
fatal cases, lost income. The researchers calculated that this cost
ranged between $10.7 and $15 million for the 188 ARI patient-study
population.
Clearly, as healthcare professionals debate the
best way to reform our healthcare system, taking steps to avoid ARIs and
these monumental treatment and societal costs should be at the top of
our list. In September, Schuetz, et al, published a study showing that
antibiotic usage can be safely avoided or minimized using a new
diagnostic tool to measure levels of procalcitonin, or PCT.7
Schuetz and his colleagues at six tertiary-care centers in Switzerland
used PCT levels to determine the etiology of lower respiratory-tract
infections (LRTIs) in more than 1,300 patients and used that information
to guide antibiotic treatment, including if and when to start treatment
and when to safely stop treatment. Prescription rates and overall
antibiotic exposure were significantly reduced in the PCT group for the
whole patient population as well as for each LRTI subgroup. The duration
of antibiotic exposure was less in the PCT group, with the overall
reduction in duration due to the PCT guidance ranging from 25.7% to
38.7% in the six study sites. The adverse effects associated with
antibiotics such as nausea, diarrhea, and rash occurred less frequently
in the PCT group.
The clinical lab is a vital partner in battling ARIs
Morton Plant Mease Health Care in Clearwater, FL,
includes four hospitals and a free-standing emergency room. At the
Morton Plant Mease critical-care department, personnel have worked
closely with clinical lab staff to form a proactive approach to find and
apply new technology and clinical practices with the goal of improving
outcomes based on a overarching commitment to enhance antibiotic
stewardship and reduce ARIs.
Physicians working closely with Morton Plant
Mease’s laboratory director have developed a program to reduce the use
of antibiotics based on the PCT test to help rule out LRTIs; suspected
sepsis; asthma and COPD flare-ups that are not caused by bacterial
infections — as well as using a sterile lavage protocol that helps rule
out contamination in cases of suspected ventilator-associated pneumonias
(VAPs).
Procalcitonin
PCT, the pro-hormone of calcitonin, was
discovered to be a sensitive biomarker for systemic bacterial infections
about 15 years ago.8 In response to bacterial infections,
nearly all tissues in the body release PCT, especially the lungs.9
In Europe, PCT is commonly used to determine a bacterial-infection
immune response from viral infections or an inflammatory response not
linked to a pathogen.10 The “SEPSIS ALERT” protocol was
started as a pilot study last year, and hospital staff is in the process
of applying the protocol to all of the Morton Plant Mease facilities
with the intent to measure the impact on patient care and antibiotic
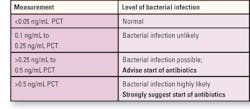
usage. In the Morton Plant Mease laboratory, the chart on page 14
indicates either the lack of or the level of bacterial infection.
Not only can the laboratory staff lead in the effort to
mitigate HAIs by pushing new policies and protocols but also by
educating its clinical colleagues.
PCT is used in the emergency department (ED) to
test those admitted patients suspected of having a significant bacterial
infection. ED protocol calls for an admitted patient with suspected
pneumonia, LRTI, or sepsis, to have three PCT tests performed in the
first 12 hours.
PCT typically spikes within the first 12 hours of
systemic bacterial infection. If the patient starts improving, we
perform the PCT test on that patient every other day. A PCT score on the
decline indicates that we are treating the patient appropriately and
that score often allows us to end antibiotic treatment once we know the
patient is safe. When PCT continues to increase over a 24-hour to
48-hour period, this is a strong indication, according to our program,
that we are not treating the patient appropriately.
Sterile lavage for suspected VAPs
The number of cases of hospital-associated
pneumonia is overwhelming, in terms of incidence, mortalities, and
treatment costs. A related area of critical concern is improving the
accuracy of cultures in patients with suspected VAP. As with PCT, a
proactive lab like that at Morton Plant Mease is vital to reducing
antibiotic overuse. When we examine patients in our hospital’s intensive
care unit (ICU) who are on ventilators and see early signs of VAP, our
standard response would be to perform a bronchial washing and send it to
the lab for culture. Invariably, these cultures were positive because
contamination from the endotracheal, or ET, tube is almost unavoidable.
The cultures show an organism that may or may not be the cause of the
infection. In fact, there may be no infection at all. This then prompts
antibiotic usage that may or may not be warranted. This problem is
common in ICUs across the country.
If the washing is taken from the lung by sending
an aliquot of saline down the endotracheal tube, then sucking it back
and culturing the specimen, the specimen is often contaminated by the
endotracheal tube colonizer.
At our hospital, we implemented sterile lavage to
get a sterile sample by passing a catheter through the endotracheal
tube within a protective catheter through the endotracheal tube within
the proactive sleeve, which we push deep into the lung. By extracting a
washing in that area under these conditions, contamination can be
avoided.
The microbiology laboratory scientist then does a
quantitative culture — a more accurate way to measure for an infection.
If we see 104 organisms per cubic centimeters per milliliter, this
indicates a serious situation which is treated aggressively. If,
however, the count is less than that, the urgency is significantly
decreased since the organism may be a contaminant. Thus, we then have
the time to monitor the patient in order to make sure that there is a
true infection before we treat.11,12
This change in the VAP protocol — designed by the
laboratory staff who were essential to its implementation — has been
successful, based on the results. Not only can the laboratory staff lead
in the effort to mitigate HAIs by pushing new policies and protocols but
also by educating its clinical colleagues. Laboratory personnel can
remind doctors and pharmacists of this at every given opportunity. We
are all partners in patient care.
By identifying resistance, the lab can help
clinicians get clear actionable information, so they can begin effective
antibiotic therapy as early as possible. The lab is critical to
monitoring resistance with surveillance campaigns of antimicrobial
resistance patterns within the hospital, and more broadly in the
community. The lab can also play the pivotal role in tracking resistance
by screening patients and healthcare workers for multidrug-resistant
organisms.
Devendra Amin, MD, F(CCP), is the medical
director of Critical Care Services at Morton Plant Hospital in
Clearwater, FL.
Note: This article is followed by another
article, “Real-time PCR testing for CDI,” that is also part of the
Continuing Education test.
References
- Centers for Disease Control and Prevention. S. aureus
and MRSA Surveillance Summary 2007.
http://www.cdc.gov/ncidod/dhqp/ar_mrsa_surveillanceFS.html .
Accessed September 21, 2009. - Science Media Centre. Experts comment on new research regarding
Community-Acquired MRSA and pneumonia, as published in
The Lancet Infectious Diseases.
http://www.sc
iacentre.org/pages/press_releases/09-05-20_lancet_camrsa.htm .
Published May 20, 2009. Accessed September 21, 2009. - Centers for Disease Control and Prevention, 2000,
NEJM. December 28, 2000. - Christ-Crain M, Jaccard-Stolz D, Bingisser R, Genday MM, et al.
Effect of PCT-guided treatment on antibiotic use and outcome in
lower respiratory tract infections: cluster-randomised
single-blinded intervention trial. Lancet. 2004;363:600-607. - Gonzales R, Malone DC, et al. Excessive antibiotic use for acute
respiratory infections in the United States. Clin Infect Dis.
2001; 33:757-762. - Roberts RR, Hota B, Ahmad I, Scott DS II, et al. Hospital and
Societal Costs of Antimicrobial Resistant Infections in a Chicago
Teaching Hospital: Implications for Antibiotic Stewardship.
Clin Infect Dis. 2009;(10).
http://www.journals.uchicago.edu/doi/abs/10.1086/605630?prevSearch=%2528Roberts%2529%2BAND%2B%255Bjournal%253A%2Bcid%255D&searchHistoryKey=.
Accessed September 23, 2009. - Schuetz P, et al. Effect of Procalcitonin-Based Guidelines vs.
Standard Guidelines on Antibiotic Use in Lower Respiratory Tract
Infections: The ProHOSP Randomized Controlled Trial. JAMA.
2009;302(10):1059-1066. - Assicot M, et al. High serum procalcitonin concentrations in
patients with sepsis and infection. Lancet. 1993;341:515-518. - Muller B, et al. Ubiquitous expression of the calcitonin-I gene
in multiple tissues in response to sepsis. J Clin Endocrinol
Metab. 2001;86:396-404. - Eberhard OK, et al. Usefulness of procalcitonin for
differentiation between activity of systemic autoimmune disease
(systemic lupus erythematosus or systemic anti-neutrophil
cytoplasmic antibody-associated vasculitis) and invasive bacterial
infection. Arthritis Rheum. 1997;40:1250-1256. - Zahar J-R, Cerf C, Maitre B, Brun-Buisson C, et al. Contribution
of Blinded, Protected Quantitative Specimens to the Diagnostic and
Therapeutic Management of Ventilator-Associated Pneumonia. Chest.
2005;128;533-544. - Guidelines for the Management of Adults with Hospital-acquired,
Ventilator associated, and Healthcare-associated Pneumonia. This
official statement of the American Thoracic Society and the
Infectious Diseases Society of America was approved by the ATS Board
of Directors, December 2004 and the IDSA Guideline Committee,
October 2004. Am J Respir Crit Care Med. 2005;171:388-416.
Real-time PCR testing for CDI improves outcomes
and reduces costs
By Brian Currie, MD, MPH
Enzyme immunoassay (EIA)
testing for toxigenic
Clostridium difficile has become standard in U.S. hospitals because of
its rapid turnaround, but the assay’s low sensitivity and specificity make
its use problematic. Since the consequences of not treating and isolating
patients with C difficile-associated infection (CDI) can be dire,
most physicians dismiss negative EIA results out of hand. As a consequence,
patients are retested, treated, and isolated unnecessarily — at great cost
to the healthcare system. Real-time polymerase chain reaction (RT-PCR)
testing for CDI provides rapid turnaround and specificity/sensitivity that
supports the elimination of most retesting and reduces the inappropriate use
of scarce hospital resources.
Clostridium difficile has become a significant
hospital-acquired pathogen, causing up to 25% of cases of
antibiotic-associated diarrhea among inpatients.1 Symptoms of CDI
range from mild diarrhea to colitis, toxic megacolon, colon perforation,
sepsis, and death.2 CDI should not be confused with non-toxigenic
or asymptomatic C difficile colonization.
C difficile produces two toxins: Toxin A, an
enterotoxin, and Toxin B, a cytotoxin. Eighty percent of toxigenic C
difficile isolates produce both toxins.3 Toxin B, produced by
virtually all toxigenic C difficile strains, is approximately 1,000
times more potent than Toxin A and is essential for disease.4
The Association for Professionals in Infection
Control and Epidemiology (APIC) estimates CDI incidence to be at least 13
per 1,000 inpatients, which is 20 times higher than previous estimates.5
According to APIC, approximately 109,000 patients die in U.S. hospitals
every year from CDI, a figure 3.5 times higher than a nearly concurrent
estimate of 28,000 deaths.6 Moreover, CDI adds between $2,454 and
$7,179 per affected patient in additional, non-reimbursable costs and up to
seven days to hospital length of stay. Estimates for costs related to CDI
treatment and prevention in the United States are in the $1 billion range,7
but based on APIC’s figures for mortality and CDI hospital patient-days this
figure is likely quite conservative.
Over the last decade, CDI epidemiology has trended
toward higher incidence, increased severity, and greater mortality. Between
1999 and 2004, deaths attributed to CDI rose from 5.7 per million population
to 23.7 per million,8 while the number of hospital discharges
with CDI more than doubled between 2001 and 2005.6 Increased
prevalence and disease severity is partly attributed to the emergence of
BI/NAP1/027, the predominant strain of C difficile in the New York
City metropolitan area. The rise of BI/NAP1/027 underscores the need to
control C difficile and exposes the shortcomings of conventional
diagnostic testing.
More than ever, rapid, accurate diagnosis of C
difficile is imperative for timely and appropriate therapy and effective
infection control.
Etiology and risk factors
C difficile is a Gram-positive anaerobic
bacillus that exists in vegetative and spore forms, and is spread through
the fecal-oral route. The spores are highly persistent and resistant to
conventional disinfection and alcohol hand-gel sanitizers, which complicates
isolation and containment strategies. When caring for CDI patients,
healthcare workers should wash their hands with soap and water instead of
alcohol-based cleansers. Soap and water does not kill spores effectively but
provides physical removal and dilution. Diluted bleach solutions are
required to disinfect patient environments.
Risk factors for CDI include antibiotic treatment,
lengthy hospital stay, age over 65 years, and severe underlying disease.9
The APIC study reported that 79% of CDI patients received antibiotics before
onset of CDI.5 Nearly all antimicrobials have been implicated in
development of CDI but cephalosporins, clindamycin, and fluoroquinolones
seem to carry higher risk.
The use of broad-spectrum antibiotics is a
potentially long-term risk factor in the etiology of CDI through the
elimination of beneficial microorganisms that compete with C difficile
in the intestinal tract and because they select for strains of C
difficile
resistant to clindamycin and fluoroquinolones.
It has been generally assumed that risk for
developing CDI diminishes in patients who discontinue antibiotic treatment.
But a recent paper found that after six weeks, intestinal flora remained
depleted in cefoperazone-treated mice, suggesting that risk may persist long
after discontinuation of therapy.10
Another study noted that C difficile spores
persist asymp-tomatically in the intestines of mice for many months with
minimal shedding of spores.11 Antibiotic treatment transformed
such mice into highly contagious “super shedders” whose digestive tracts
were depleted of beneficial bacteria. Lacking normal digestive flora, these
mice experienced overgrowth of C difficile and excreted high levels
of infectious spores.
These results hearken back to a small human study on
patients with recurrent C difficile colitis who had received up to
seven courses of systemic antibiotics. After normal intestinal flora were
re-introduced, only one of 16 evaluable patients experienced a relapse of
C difficile colitis.12
Diagnosis
A positive diagnosis of CDI requires that the patient
be symptomatic and test positive for toxigenic
C difficile, or have a pathologic colon specimen consistent with
pseudomembraneous colitis, or show evidence of pseudomembraneous colitis on
colonoscopy.13 When diarrhea is the primary symptom, three or
more episodes per day over one to two days may be a reasonable trigger for
ordering a diagnostic test for toxigenic C difficile.14
Following such rules could reduce unnecessary testing by up to 39%.15
Diarrhea can have many etiologies, however, and is not always easy to
characterize clinically.
A good working definition of diarrhea is any stool
that takes the shape of its container. C difficile testing,
therefore, should be restricted to such samples. Simply asking about the
frequency of loose stools they are experiencing can help screen patients at
high risk for CDI and reduce unnecessary testing.14
Conventional diagnostic tests for CDI lack an
acceptable combination of sensitivity, specificity, and timeliness. Stool
culture, the most sensitive test, is labor-intensive, takes several days,
and does not differentiate between toxigenic and non-toxigenic C
difficile
strains. Thus, confirmation of CDI requires an additional toxin-detection
test which adds time and cost.
The cytotoxicity assay measures the production of
Toxin B and the cytopathologic effect of a stool-sample preparation on
cultured cells.16 Although it is sometimes considered the “gold
standard” for detection of toxigenic C difficile and CDI diagnosis,
it is less sensitive than toxigenic culture.17 In addition,
cytoxicity assays are expensive, require extensive operator input, and take
three to seven days.
Enzyme immunoassays use antibodies to one or both
C difficile toxins. EIAs are inexpensive and take less than four hours,
and have, therefore, become the default hospital-based test for CDI.
Unfortunately, Toxin A/B EIAs have poor sensitivity (50% to 99%) and
specificity (70% to 100%).18 Clinicians often treat regardless of
the EIA result, which leads to over-treatment and unnecessary isolation.19
Another rapid assay, for glutamate dehydrogenase
(GDH), a “common antigen,” is based on EIA as well. Older versions of this
test used latex agglutination, which is less sensitive. The GDH assay alone
does not distinguish toxigenic from non-toxigenic strains of C difficile,
and, thus, requires a second confirmatory test for the toxigenic pathogen.
The GDH assay’s high sensitivity and negative predictive value (NPV) make it
somewhat useful as a screening test for C difficile, but not for CDI.20
GDH testing returns almost all true-positive results and some
false-positives.16 But when coupled with cytotoxicity testing to
differentiate false-positives from true-positives, the sensitivity of the
two-step algorithm for detection of toxigenic C difficile fell to
77%.21 Thus, this two-step assay cannot be recommended for CDI
testing.16
Real-time PCR
Polymerase chain reaction (PCR) assays represent the
most significant recent development in CDI testing. Accurate, experimental
PCR amplification methods for C difficile have existed since at least
1952 but are unsuitable for real-time testing because they require pre-test
DNA purification steps which are hard to standardize and are not FDA
certified.
At Monefiore Medical Center, we have had considerable
experience with an assay which amplifies tcdB, the gene coding for C
difficile Toxin B.4 The assay employs quantitative real-time
PCR (qPCR), which simultaneously amplifies and detects the gene
target thereby saving several hours compared conventional PCR. The test
takes approximately one hour, involves 15 minutes of operator time, and runs
on PCR equipment found in most labs. In addition, the assay has been
optimized to allow direct stool swab sampling. The test’s sensitivity and
specificity of more than 95%.23 User groups have reported
sensitivities for this assay ranging from 94% to 100%,24,25 with
NPVs of 99% and greater.24,25 High NPVs permit clinicians to rule
out a diagnosis of CDI from negative tests with confidence, a factor with
great implications for patient health and hospital finances.
Comparison of methods
When using Toxin A/B EIA testing for C difficile,
physicians often compensate by retesting patients with negative test results
and by overtreating suspected cases. While national standards discourage
retesting, few laboratories enforce these guidelines, and physicians
generally ignore them.
Both positive and negative EIA results carry
independent significance but only to the extent that the test result is
accurate. Positive results prompt initiation of therapy and
infection-control interventions; a negative result contraindicates treatment
and isolation for CDI, while suggesting a workup for an alternative
etiology.
False-positive and false-negative EIA test results
also carry significant implications. Based on known statistics, EIA testing
of symptomatic patients could be expected to return approximately 2%
false-positives and 10% false-negatives.26 Thus, for every 1,000
patients tested, approximately 20 will experience hospital stays prolonged
by up to seven days, usually in isolation, and receive a 10-day course of
antibiotics. Conversely, 100 patients who actually have CDI will not be
isolated or treated.
Isolating patients is expensive. Personal protective
equipment costs approximately $2 per patient visit, which adds up quickly
when meals, examinations, and cleaning visits are considered. Accurate
diagnostic testing could virtually eliminate inappropriate isolation, thus
freeing hospital resources for more urgent caregiving.
Of the 85% or so of patients with suspected CDI whose
tests are negative by Toxin A/B EIA, 10% to 12% (about 100 patients) turn
out to be false by cytotoxicity testing. Theoretically, false-negatives
would not be treated. In reality, clinicians using Toxin A/B EIA testing
treat most patients with a negative test anyway. In our hospital — Monefiore
Medical Center in the north Bronx, NY — close to 40% of those patients
undergo a full, 10-day CDI antibiotic course, which for vancomycin carries a
price tag of $168. Direct costs associated with EIA retesting can also be
significant. Most negatives with diarrhea are retested — some, numerous
times — and nearly all are isolated.
A rapid, reliable assay like qPCR reduces
false-positives by half. Even greater benefit accrues from the
near-elimination of false-negatives, to about 1% of patients tested, from
10% to 12% for EIA. A near-zero rate of false-negatives introduces, for the
first time in the management of CDI, a scientific basis for appropriate
diagnosis, treatment, infection-control interventions, and the elimination
of retesting.
Available studies suggest that qPCR assays for
the Toxin B gene (tcdB) are the most sensitive, specific and
appropriate tests for confirming a CDI diagnosis, but some experts have
argued that qPCR are too sensitive and that false-positives would
arise from its ability to detect colonizing C difficile, which does
not cause disease.16
This argument should apply to all tests for toxigenic C difficile;
but, in fact, no study has demonstrated that a more sensitive test is more
likely to detect colonizing bacteria. The best solution to this concern is
to restrict testing to patients who are likely to have CDI, that is those
who meet the criterion of frequent episodes of diarrhea.16
Conclusion
q PCR provides both high sensitivity and rapid
turnaround time, factors that could potentially revolutionize treatment of
C difficile infection and transmission. While qPCR is more
expensive than EIA (about $25 vs. $8), improved diagnostic testing provides
significant opportunities to reduce CDI-related treatment and isolation
costs. Therefore, qPCR should be considered as a cost-effective
option.
The performance of qPCR supports laboratory
policies that severely limit repeat testing. Realizing the cost-saving
potential, however, will require significant caregiver education on the
capabilities of qPCR to guide patient management. Monefiore Medical
Center is in the process of linking qPCR results to an Antibiotic
Stewardship Program to accomplish these goals and to achieve optimal
savings.
Brian Currie, MD, MPH, is vice president and medical
director for research at Monefiore Medical Center in the north Bronx, NY,
which uses the BD GeneOhm Cdiff assay for testing for C difficille
Toxin B. Dr. Currie is also assistant dean for clinical research at the
Albert Einstein College of Medicine, a graduate school of Yeshiva University
in Morris Park, also in the Bronx.
References
- Blossom DB, McDonald LC, et al. The challenges posed by reemerging
Clostridium difficile infections. Clin Inf Dis.
2007;45:222-227. - Centers for Disease Control and Prevention. Information for
healthcare providers.
http://www.cdc.gov/ncidod/dhqp/id_CdiffFAQ_HCP.html . Published
July 22, 2005. Accessed June 27, 2008. - McFarland LV, Beneda HW, Clarridge JE, Raugi GJ. Implications of the
changing face of Clostridium difficile
disease for health care practitioners. Am J of Infect Control.
2007;35(4):237-253. - Lyras D, et al. Toxin B is essential for virulence of
Clostridium difficile. Nature. 2009;458(4):1176-1179. - Jarvis WR, Schlosser J, Jarvis AA, Chinn RY, et al. National point
prevalence of Clostridium difficile in US health care facility
inpatients. Am J Infect Control. 2009;37(4):263-270. - McDonald LC. The changing epidemiology of
Clostridium difficile. Poster session presented at: June 2008 Annual
Meeting of the Association for Professionals in Infection Control and
Epidemiology, Denver, CO. - Dubberke ER, Reske KA, Butler AM, et al. Attributable outcomes of
endemic Clostridium difficile-associated disease in non-surgical
patients. Emerg Inf Dis. 2008;14:1031. - Redelings MD, Sorvillo F, Mascola L. Increase in
Clostridium difficile-related mortality rates, United States,
1999-2004. Emerg Infect Dis.
http://www.cdc.gov/EID/content/13/9/1417.htm . Published
September 2007. Accessed September 14, 2009. - Sunenshine RH, McDonald LC. Clostridium difficile-associated
disease: new challenges from an established pathogen. Cleve Clin J
Med. 2006;73:187-197. - Antonopoulos DA, Huse SM, Morrison HG, et al. Reproducible Community
Dynamics of the Gastrointestinal Microbiota following Antibiotic
Perturbation. Infection and Immunity. 2009;77(6):2367-2375. - Lawley TD,Clare S, Walker AW. Antibiotic Treatment of
Clostridium difficile Carrier Mice Triggers a Supershedder State,
Spore-Mediated Transmission, and Severe Disease in Immunocompromised
Hosts. Infection and Immunity. 2009;77(9):3661-3669. - Aas J, Gessert CE, Bakken JS. Recurrent
Clostridium difficile colitis: case series involving 18 patients
treated with donor stool administered via a nasogastric tube. Clin
Infect Dis. 2003;36(5):580-585. - McDonald et al. Recommendations for Surveillance of
Clostridium difficile-Associated Disease. Infect Control Hosp
Epidemiol. 2007; 28:140-145. - Peterson LR, Robicsek A. Does My Patient Have
Clostridium difficile Infection? Annals of Int Med.
2009;151:176-179. - Katz DA, Lynch ME, Littenberg B. Clinical prediction rules to
optimize cytotoxin testing for Clostridium difficile in
hospitalized patients with diarrhea. Am J Med. 1996; 100:487-495. - Alcal’a L, S’anchez-Cambronero L., Catal’an MP, et al. Comparison of
Three Commercial Methods for Rapid Detection of
Clostridium difficile Toxins A and B from Fecal Specimens. J Clin
Microbiol. 2008;46(11): 3833-3835. - Peterson LR, Manson RU, Paule SM, et al. Detection of Toxigenic
Clostridium difficile in Stool Samples by Real-Time Polymerase Chain
Reaction for the Diagnosis of C difficile-Associated Diarrhea.
Clin Infect Dis. 2007;45:1152. - Fedorko DP, Engler ED, O’Shaughnessy EM, et al. Evaluation of two
rapid assays for detection of Clostridium difficile
toxin A in stool specimens. J Clin Microbiol.1999;37:3044-3047. - Morelli MS, Rouster SD, Gianella RA, et al. Clinical Application of
Polymerase Chain Reaction to Diagnose Clostridium difficile in
Hospitalized Patients With Diarrhea. Clin Gastroenterol Hepatol.
2004;2:669-674. - Fenner L, et al. Rapid and Reliable Diagnostic Algorithm for
Detection of Clostridium difficile. J Clin Microbiol.
2008;46(1):328-330. - Reller ME, Lema CA, Perl TN, et al. Yield of Stool Culture with
Isolate Toxin Testing versus a Two-Step Algorithm Including Stool Toxin
Testing for Detection of Toxigenic Clostridium difficile. J Clin
Microbiol. 2007;45(11):3601-3605. - Arzese A., Trani G, Riul L, Botta GA. Rapid polymerase chain
reaction method for specific detection of toxigenic
Clostridium difficile. Eur J Clin Microbiol Infect Dis.
1995;14:716-719. - Metzger B. Comparison of BD GeneOhm Cdiff PCR Assay and the Meridian
Premier Toxins A&B for Detection of C difficile
from Clinical Specimens Poster session presented at: March 2009 Annual
Meeting of the Society for Healthcare Epidemiologists of America, San
Diego, CA. - Alcabasa R, Aird D, Wehrlin J, et al. Comparison of the BD GeneOhm
Cdiff Assay (BD GeneOhm, San Diego, CA) to the Wampole
Clostridium difficile Toxin B Test (TechLab, Blacksburg, VA). Poster
session presented at: 2008 annual meeting of the American Society for
Microbiology, Boston, MA. - Fuller D, Buckner R, Newcomer K, et al. Clinical Comparison of the
Molecular-Based BD GeneOhm Cdiff Assay to the Cytotoxicity Tissue
Culture Assay for the Direct Detection of Toxin B gene from Toxigenic
Clostridium difficile Strains in Fecal Specimens. Poster session
presented at: 2008 Annual Meeting of the Anaerobe Society of the
Americas, Long Beach, CA. - Other data (Currie B. Unpublished data, 2009).