The challenge of antimicrobial resistance for the clinical laboratory: The role of the antibiogram
To take the test online go HERE. For more information, visit the Continuing Education tab.
LEARNING OBJECTIVES
Upon completion of this article, the reader will be able to:
1. List the reasons that have led to antimicrobial resistance.
2. Discuss healthcare statistics and outcomes of multidrug-resistant bacteria.
3. Describe how proper antibiotic treatment is selected for individual patients.
4. Discuss how antibiograms are generated, used, and developed.
Simply stated, antimicrobial agents, which include antibacterial, antifungal, antiviral, and antiparasitic agents, are life-saving medicines that are losing their effectiveness globally. A combination of over prescribing and indiscriminate use in human, veterinary, agriculture and other sectors, coupled with the continuing evolution of novel resistance mechanisms, has led to a pandemic of resistant organisms globally. Unlike COVID-19, this pandemic has been escalating for several decades, causing some to refer to it as “the hidden threat.”1 For many patients with life threatening infections that resist even our newest antimicrobial agents, it is all too real. Multidrug-resistant bacteria have emerged not only in healthcare settings, but also in our food supply, and even in our pets. Among humans living in the United States, data from the Centers for Disease Control and Prevention show that a new antimicrobial resistant infection develops every 11 seconds and every 15 minutes someone dies of an antimicrobial resistant infection. That translates into 2.8 million new resistant infections in the United States every year and >35,000 deaths.2 Globally, the data are much starker with deaths associated with antimicrobial-resistant organisms approaching 5 million annually.3 Infections caused by antimicrobial-resistant organisms lead to longer hospital stays, more serious complications, and increased mortality rates because effective treatment is delayed or simply not available. Everyone, not just immune compromised patients, is at risk.
How did we get to this point? For many years, there was a sense among physicians that antimicrobial agents, even if they were not really needed, caused no harm. In other words, many antibiotics were prescribed for patients “just to be on the safe side.” Thus, the barrier to prescribing antimicrobial agents, especially for respiratory illnesses in both children and adults, was low. We now know better. The adverse drug events that can occur can send people to emergency rooms with nausea, vomiting, skin rashes, and in some instances anaphylaxis. Yet, a more insidious side effect is infection with an anaerobic toxin-producing gram-positive bacillus known as Clostridioides (formerly Clostridium) difficile, or more simply “C. diff.” When antimicrobial agents disrupt the bowel flora, many of the resident microorganisms that form a natural barrier to infection with pathogenic microbes are killed. This allows colonization of the gut with C. diff that leads to infection. Infection with C. diff is associated with elaboration of one or more toxins, resulting in gastrointestinal disease that can range from mild diarrhea to pseudomembranous colitis and even death.4 More than 225,000 C. difficile cases are observed in the US annually according to the CDC.2 In fact, we are still learning about the long term-side effects of antimicrobial use including effects on immune function and metabolism.5 Thus, taking an antimicrobial agent is not without risk and should be reserved for treating infections, where the benefits of the taking the drug clearly outweigh the risk of adverse events. Even so, a World Health Organization (WHO) study in 2020 among 2000 COVID-19 cases from multiple countries, reported that 72% of patients received antimicrobial agents even though only 8% had a documented bacterial or fungal infection.6 Granted, early on in the COVID-19 pandemic, we didn’t know what the risk of secondary bacterial infections was so antimicrobial agents flowed freely to patients. But, even as our understanding of COVID-19 infections expanded and secondary bacterial infections were noted to occur in <10% of cases, antimicrobial agents still flowed freely. This is but one example of how the overuse of antimicrobial agents can fuel the development of resistant microorganisms.
Before the onset of COVID-19, the combined efforts of public health organizations, physicians, laboratorians, and professional societies in the United States were making progress in lowering the numbers of healthcare-associated infections, such as methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), and drug-resistant Pseudomonas aeruginosa.7 However, that progress was reversed with resurgences of these organisms in hospitals during COVID-19 when our concern for transmission of COVID-19 among patients took priority over transmission of traditional healthcare–associated pathogens, including MRSA and VRE. There was one notable exception to the successful reduction of healthcare-associated infections in the United States prior to COVID-19: the rates of infections with carbapenem-resistant gram-negative organisms remained stable. Carbapenems are often referred to as the antibiotics of last resort and the development and spread of organisms that are resistant to this class of agents has been cited as a global menace.8 Outside of the hospital, two other antimicrobial resistance problems were gaining momentum. In 2020, rates of infections caused by Mycobacterium tuberculosis (the bacterial species that causes tuberculosis) increased for the first time in over a decade. Years of progress in fighting tuberculosis were reversed in a single year due to COVID-19’s impact on tuberculosis control programs, which were often halted to deal with the COVID-19 pandemic. Not only M. tuberculosis strains but multidrug-resistant M. tuberculosis strains were on the increase, which was a major blow to public health programs globally. Besides tuberculosis control programs, the other public health efforts that were extremely compromised by COVID-19 were those focused on preventing sexually transmitted infections. Multidrug-resistant strains of the sexually transmitted bacterial pathogen Neisseria gonorrhoeae emerged.9 The challenges of resistance are growing both inside and outside of hospitals worldwide.
The laboratory perspective
What is often overlooked when considering all the journal articles and news stories on antimicrobial resistance are the Herculean efforts of the clinical laboratory, and the microbiology laboratory in particular, to generate all of the data on the emergence of resistant microorganisms in both hospital and community settings. The microbiology laboratory remains the unsung hero in the fight to control the spread of resistance globally. Without the antimicrobial susceptibility data that flows from the microbiology laboratory, physicians would not know how to select the most effective therapy for patients with infections, infection preventionists would not know which patients with resistant organisms needed to be placed in contact precautions to prevent spread in hospitals, and public health officials would not know where outbreaks of multidrug-resistant infections were occurring.
Guiding treatment for the individual patient
Physicians, pharmacists, and antimicrobial stewardship committees look to the microbiology laboratory to guide the selection of antimicrobial agents via antimicrobial susceptibility test results to treat patients with infections. To perform this function, the laboratory first isolates the microbial pathogen from clinical specimens (e.g., blood, urine, sputum, or wounds), identifies the bacterial species, and generates the antimicrobial susceptibility pattern of the isolate to a variety of antimicrobial agents via some form of broth microdilution or disk diffusion testing. This traditional approach may take up to 72 hours to complete, which for critically ill patients may be too long to impact care. Thus, rapid molecular diagnostic methods can be employed to detect organisms directly in clinical specimens (such as sputum or wounds) or from positive blood culture specimens, reducing the turnaround time of identification sometimes to as little as one hour. Some commercial panels also have a limited selection of antimicrobial resistance genes that can be used to predict resistance to some classes of antimicrobial agents for bacterial pathogens earlier in the course of infection. A meta-analysis of the impact of molecular methods shows that using these rapid laboratory tests significantly improves patient outcomes, especially when the results were shared with an antimicrobial stewardship committee.10
Generating antibiograms
Traditional susceptibility test results guide the treatment of the individual patient, but when the results from all patient specimens over the course of a year are pooled, the cumulative percent susceptibility data for the most commonly used antimicrobial agents for the key bacterial pathogens enables physicians to formulate anti-infective strategies before the data from the standardized susceptibility tests are known. This is known as “empiric therapy” and the cumulative susceptibility tests results are known collectively as the hospital’s “antibiogram.” These results are critical for the management of patients in the early stages of infection. For septic shock, every hour a patient is on ineffective therapy, the chance of death increases by 7.6%.11 Thus, the antibiogram is relied upon to narrow therapeutic choices to those most likely to be effective. When additional data become available through standard susceptibility testing or molecular test methods, the therapy is adjusted appropriately.
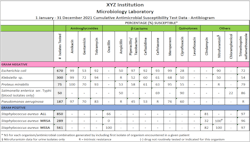
Antibiograms are typically assembled at least annually, often in collaboration with the hospital’s pharmacy and the antimicrobial stewardship team. The report typically includes data for both gram-positive and gram-negative organisms (See Figure 1). Laboratories often customize their antibiograms to provide more detailed information on specific patient populations or hospital units. For example, a separate antibiogram may be generated for outpatients, for pediatric patients, or for hematology/oncology patients. Further stratification may be done for blood and urine isolates depending on the hospital’s needs. The Clinical and Laboratory Standards Institute has produced document M39, which focuses on guiding laboratories in the preparation of their antibiograms.12
Antibiograms not only help guide physicians and pharmacists in selecting the best empiric antimicrobial treatment while culture and susceptibility results are pending, antibiograms also impact the broader healthcare ecosystem by providing data that can guide infection prevention programs designed to contain the spread of antimicrobial-resistant infections in the hospital. Following the annual incidences of MRSA, VRE, and carbapenemase-producing organisms (CPO) through antibiograms is one indicator of the effectiveness of infection prevention programs in a hospital. However, the data, if getting worse, may be signaling the emergence of new multidrug-resistant strains or organisms with novel resistance mechanisms. In some cases, it may be of public health value to share resistance data more broadly, thus, the aggregated susceptibility data from a region can be exported to external surveillance systems and used to understand the epidemiologic spread of resistant organisms. Tracking CPOs has become a major public health priority. This is because serine-based carbapenemases, like KPC, can often be treated with one of the newer beta-lactam/beta-lactamase inhibitor combinations (ceftazidime/avibactam, meropenem/vaborbactam, or imipenem/relebactam) while organisms with metallo-carbapenemases, like NDM, IMP, and VIM, typically do not respond to these new agents. This is very important information especially for those committees developing empiric therapy guidelines.
Among the many challenges for the microbiology laboratory when populating antibiograms with data is making sure that they are using the most current interpretive criteria, i.e., susceptible, intermediate, and resistant interpretations (also known as breakpoints). Laboratories in the United States adhere to the standards produced by the Clinical and Laboratory Standards Institute. The definitions of susceptibility and resistance are fluid and change over time with the accumulation of clinical use and outcomes data and the emergence of new resistant strains. Both minimal inhibitory concentration (MIC) and disk diffusion criteria may change. Significant changes include, but are not limited to, the interpretations of cephalosporins, carbapenems, and fluoroquinolones. It often takes a few years for the automated susceptibility testing systems to incorporate the new interpretive criteria in the instrument’s software once the updated criteria are approved by the U.S. Food and Drug Administration. When released, the laboratory needs to verify the updated criteria in-house. A recent review showed that many laboratories in the United States have yet to incorporate new breakpoints especially for cephalosporins and carbapenems and this may lead to misidentifying resistant organisms as drug susceptible.13 That in turn may lead to treatment failures and poor patient outcomes. If the outdated criteria are incorporated into antibiograms, the impact is compounded. Thus, it is incumbent on the microbiology lab to make sure it is using the most up-to-date criteria to ensure that positive patient outcomes are realized. In fact, using updated breakpoints is now a requirement of the College of American Pathologists.14
Aids for developing antibiograms
There are multiple sources of information to aid laboratories in preparing their antibiograms. In addition to CLSI document M39 (which must be purchased), a free software program called WHONET (https://whonet.org/) is available that can download data from automated susceptibility testing methods or import disk diffusion data from spread sheets. Data can be displayed in a variety of ways for the laboratory, the antimicrobial stewardship committee, and for infection control practitioners. The program also facilitates exporting data to surveillance systems, such as the World Health Organization GLASS program.
Summary
Antimicrobial resistance is a global problem. When antibacterial agents are used inappropriately, resistance develops in bacterial strains and patients’ therapies fail. The laboratory plays a central role in providing the antimicrobial susceptibility test results that guide treatment of individual patients and, when assembled in an annual antibiogram, guides empiric therapy, infection prevention, and antimicrobial stewardship activities. It is important to ensure that the susceptibility data generated are reported with the latest interpretive criteria. Antibiograms are an important tool to inform infection prevention activities and monitor resistance trends over time. Thus, antibiograms are a critical factor in our efforts to bring antimicrobial resistance under control.
REFERENCES
- Gerberding JL. Antibiotic resistance: The hidden threat lurking behind Covid-19. STAT. Published March 23, 2020. Accessed December 5, 2022. https://www.statnews.com/2020/03/23/antibiotic-resistance-hidden-threat-lurking-behind-covid-19/.
- CDC. Antibiotic Resistance Threats in the United States, 2019. Centers for Disease Control and Prevention; 2019. Published December 2019. Accessed December 5, 2022. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;12;399(10325):629-655. doi: 10.1016/S0140-6736(21)02724-0.
- Martin JS, Monaghan TM, Wilcox MH. Clostridium difficile infection: Epidemiology, diagnosis and understanding transmission. Nat Rev Gastroenterol Hepatol. 2016;13(4):206-16. doi: 10.1038/nrgastro.2016.25.
- Bejaoui S, Poulsen M. The impact of early life antibiotic use on atopic and metabolic disorders: Meta-analyses of recent insights. Evol Med Public Health. 2020 Oct 24;2020(1):279-289. doi: 10.1093/emph/eoaa039.
- Maragia M. Covid-19 is driving up antibiotic resistance. The Standard. Published November 27, 2020. Accessed December 5, 2022. https://www.standardmedia.co.ke/commentary/article/2001395337/covid-19-is-driving-up-antibiotic-resistance.
- CDC. The biggest antibiotic-resistant threats in the U.S. Centers for Disease Control and Prevention. Published July 15, 2022. Accessed December 5, 2022. https://www.cdc.gov/DrugResistance/Biggest-Threats.html.
- Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J Infect Dis. 2017;15;215(suppl_1):S28-S36. doi: 10.1093/infdis/jiw282.
- Wi T, Lahra MM, Ndowa F, Bala M, Dillon JR, Ramon-Pardo P, et al. Antimicrobial resistance in Neisseria gonorrhoeae: Global surveillance and a call for international collaborative action. PLoS Med. 2017;7;14(7):e1002344. doi: 10.1371/journal.pmed.1002344.
- Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: A systematic review and meta-analysis. Clin Infect Dis. 2017 Jan 1;64(1):15-23. doi: 10.1093/cid/ciw649.
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589-96. doi: 10.1097/01.CCM.0000217961.75225.E9.
- Clinical and Laboratory Standards Institute. Analysis and presentation of cumulative antimicrobial susceptibility test data; Approved Guideline - Fifth Edition. M39-A5. Clinical and Laboratory Standards Institute, Wayne, PA; 2022.
- Simner PJ, Rauch CA, Martin IW, Sullivan KV, Rhoads D, Rolf R, et al. Raising the bar: Improving antimicrobial resistance detection by clinical laboratories by ensuring use of current breakpoints. Open Forum Infect Dis. 2022;7;9(3):ofac007. doi: 10.1093/ofid/ofac007.
- Updating breakpoints in antimicrobial susceptibility testing. Asm.org. Published February 22, 2022. Accessed December 5, 2022. https://asm.org/Articles/2022/February/Updating-Breakpoints-in-Antimicrobial-Susceptibili#:~:text=The%20CAP%20checklist%20update%20(found,2021%20(at%20a%20minimum).
About the Author

Fred C. Tenover, Ph.D., D(ABMM), FIDSA, FAAM, FISAC
is Vice President for Scientific Affairs at Cepheid, where he has worked since 2008. He is also Consulting Professor of Pathology at Stanford University School of Medicine, Adjunct Professor of Epidemiology in the Rollins School of Public Health at Emory University, and Consulting Professor of Biology at the University of Dayton. Prior to joining Cepheid, he served for 18 years at the Centers for Disease Control and Prevention (CDC) in Atlanta as Associate Director for Laboratory Science in the Division of Healthcare Quality Promotion. His research team discovered the KPC carbapenem resistance gene and other key resistance determinants. He then became the Director of the Office of Antimicrobial Resistance for the CDC. Dr. Tenover was a member of the CLSI Antimicrobial Susceptibility Testing Subcommittee for 17 years, developing novel test methods for resistance detection and interpretive criteria for new antimicrobial agents. He is a Diplomate of the American Board of Medical Microbiology and a Fellow of the American Academy of Microbiology, the Infectious Disease Society of America, and International Society of Antimicrobial Chemotherapy.