It’s all in the genes—expanding molecular testing to improve healthcare for those with blood disorders
Earning CEUs:
Information the genes impart on man’s characteristics including future disease conditions has not been always understood. Friedrich Miecher, a Swiss Chemist, was the first to expand man’s knowledge of nucleic acid in the 1860’s; however, Watson and Crick greatly improved our understanding of deoxyribonucleic acid (DNA) over 60 years ago. Since then the genes that control the sequence of DNA and the role of ribonucleic acid (RNA), as well as the products of the molecular codes, continues to be clarified. Molecular testing has moved from the research laboratory to the clinical laboratory. This transition has enabled science to gain insight into genetic differences, mutations (ie: Huntington Disease, Down’s syndrome, Philadelphia chromosome), diseases, cell growth, cancer onset, and blood disorders; moving medical science to a new frontier.
The part of the gene that codes for amino acids responsible for protein structure is referred to as an exon. These protein products are biomarkers which can appear in the plasma or on cell surface to indicate normal or abnormal cellular function. Expanding our understanding of these biomarkers has been extremely beneficial in diagnosis and management of various disease states and avoiding adverse events.
Modern medicine has harnessed molecularly determined biomarkers to manage various disease states. The recent 2018 Nobel Prize award in Physiology or Medicine highlighted this. Drs James P. Allison and Tasuku Honjo were honored for advancing cancer treatment by using the host’s immune system to arrest or limit cancer. Honjo’s discovery of Program Cell Death-1 (PD-1) has been effective in treating lung cancer, renal cancer, lymphoma, and melanoma.
Blood groups are biodeterminants
So, what about using molecular testing for blood disorders? In the clinical laboratory, the blood bank has been at the forefront of routinely testing for biomarker determinants either through the antigens formed by genetic uniqueness of the patient or the immune response to those biomarker determinants. In the early 1900s, Karl Landsteiner was the first to demonstrate the phenotypic differences of the blood group system, later known as the ABO blood group system. The ABO system continues to be the most important blood group system with antigens expressed on red cells, platelets, and other tissues. At the time of Landsteiner’s description of the ABO system, he did not know that these observed differences were gene-controlled glycoproteins with specific carbohydrates (sugars) forming immunodominant carbohydrate epitopes. The ABO system is controlled by the ABO gene and another precursor gene (H gene), responsible for the H antigen. When the H antigen is present, and depending on the presence or absence of A and B glycotransferase, N-acetylgalactosamine (GalNAc) will form the A antigen or Galactose (Gal) will form the B antigen or no additional sugar will be added to the H antigen for a phenotype of group O. If the H antigen is absent, the red cells are of the very rare Bombay phenotype.
Although the mechanism is not fully understood, there appears to be a relationship between the ABO and other carbohydrate blood group systems with malignancy, cellular adhesion, and some infectious disease.1,2 West Nile Virus and Zika Virus have been associated with the red cells and may adhere throughout the erythrocytes’ lifespan.3,4
Next to the ABO system, the Rh system, and the Kell system, are blood group systems of clinical significance in transfusion practice. In 1939, Levine and Stetson observed that the serum of a pregnant woman agglutinated 80 percent of ABO compatible samples, resulting in the identification of the Rh blood group system. The Kell system was the first system identified following Coombs technique, which uses anti-human globulin to detect antibodies adhered to the red cells. To date, the field of immunohematology has identified 54 antigens associated with the Rh system and 36 antigens in the Kell system. Unfortunately, many of these discoveries have been associated with adverse events in women of child bearing age and those individuals with chronic transfusion dependent diseases.
What is observed is not necessarily what is encoded in the genes. The phenotype is the observed characteristics of the gene. In the clinical immunohematology laboratory, the observed characteristics (phenotype), based on antibodies formed by an individual lacking an antigen, can be different than expressed by the gene. Antigens can be a mosaic of determinants or epitopes influenced by other exons. The genotype represents the genetic makeup, that is, the true expression of the genes inherited by the individual.
Alloimmunization in chronically transfused individuals
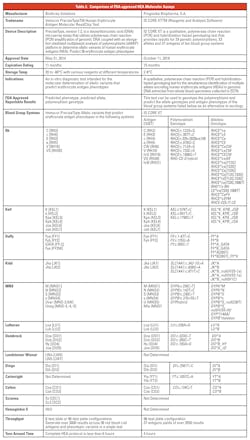
The risk of alloimmunization is a major concern for those dependent on transfusion therapy. Table 1, “Rates of Alloimmunization in Transfused Recipients” is a summary of those alloimmunization risks in both the occasionally transfused and those with blood disorders dependent on transfusions. Erythrocyte alloimmunization occurrence ranges from 4.4 percent to 76 percent with a higher frequency of antibodies to antigens of the Rh system.5 Age, gender (female), and number of transfusion (red blood cells) were major risk factors; however other risk factors were identified.Alloimmunization in blood disorders requires additional time in identification and delays treatment, all contributing to healthcare costs. Extended phenotypically matching blood has been used for many years but should be performed prior to transfusions and the formation of antibodies. For example, if an individual has been transfused, phenotyping may also detect a mixed field of transfused cells depending on if the transfused cells are still circulating in the recipient. This mixed red cell population would render the patient’s own blood type indeterminable. For recently transfused patients, special procedures are required to separate the individual’s own cells from those transfused. Blood group genotyping (BGG) does not have these limitations since the individual with a blood disorder can provide DNA anytime to determine the genotype of the red cell antigens.
Sickle cell disease (SCD) individuals are dependent on transfusions and many times demonstrate multiple antibodies to red cell antigens. Gehrie et al. point out that the greatest adverse events associated with transfusions to SCD individuals is the delayed hemolytic transfusion reactions, which can be life threatening.6 Ten years ago the cost of treating an individual with SCD was about $460,000.7 Since 2009, the U.S. national health spending has increased from $2,495.4 billion to $3,337.2 billion in 2016; a 25.2 percent increase. While current data is not available specifically for SCD, it can be assumed that SCD treatment and transfusion medicine workups have also increased by a comparable rate. Campbell et al. recommends hemoglobin S negative, prophylactically matched antigens for Rh and Kell in SCD patients.8 Not surprisingly, extra attention to extended phenotypical matched antigens have also been effective in those general surgery patients by reducing their alloimmunization rate by 65 percent.9
Cost effectiveness of alloimmunization prevention
Prevention of alloimmunization in blood disorders such as SCD, thalassemia, and myelodysplasia syndrome is costly. Cost of serological testing and ordering specific blood is expensive with the 2019 CMS Outpatient Prospective Payment reimbursement set at $32.89 per Rh(d) phenotype and $271.73 for donor antigen typing. In addition, success with serologic matching of antigens, especially in African Americans is variable based on high frequency of variant Rh alleles in the SCD population. On the other hand, a genotypic matching program could offer cost savings since the recipient and donor are only genotyped once. Many blood centers have initiated genotyping and maintaining on a set number of reliable donors with genotype status in their available donor pool. For example, the French have developed a strategy for providing genotypically matched blood to “high-responders” (increase alloimmunization) and newly diagnosed SCD patients.10 In the U.S., the National Institutes for Health (NIH), and the Wellcome Trust have funded the MedSeq Project. (The MedSeq Project, funded by the NIH, was the very first study exploring the use of whole genome sequencing (GS) in both a healthy population and a population with suspected genetic cardiac disease.) According to Lane et al., “By enabling more precise antigen-matching of patients with blood donors, antigen typing based on whole-genome sequencing provides a novel approach to improve transfusion outcomes with the potential to transform the practice of transfusion medicine.”11
BGG has been strongly advocated to determine appropriate Rh immune globulin therapy (RhIg) to prevent the formation of alloimmunization to anti-D. RhIg is a biological product extremely effective in prevention of hemolytic disease of the fetus and newborn (HDFN) due to anti-D alloimmunization. The D antigen is mosaic of epitopes and if one part of the mosaic is missing the observed laboratory characteristic may be interpreted as “weak.” In the U.S., there is a lack of standardization to workup and interpret a weak D. In many medical facilities, a woman serologically phenotyped as a weak D, and if she were of child bearing age, would be managed as a D negative and would receive RhIg therapy. A weak D is considered to be a non-reactive or less than a 2+ reaction, usually detected only by 37º C incubation and anti-D binding with anti-human globulin. Sandler et al. advocates that BGG to confirm the D status would avoid at least 24,700 unnecessary ante- and postpartum RhIG injections.12 In 2018, the mini-dose used for ante partum treatment was covered at $34.74 and $95.39 for the postpartum RhIG injection; genotyping would save from $858,078 to $2,356,133 in cost avoidance from RhIG injections. Furthermore, Rh negative blood is overused, especially in those with a serologic weak D phenotype. If the genotype were determined, approximately 48,000 units of RhD negative blood could be available for emergency cases when RhD negative blood is needed for mass transfusion protocols. Sandler et al. advocates the expansion of genomic testing to personalize
medical care.
Multiple myeloma is a blood cancer of malignant plasma cells in the bone marrow. A monoclonal antibody (anti-CD38) also known as Darzalex (daratumumab) has recently been used for treatment of multiple myeloma in which other lines of therapy have been ineffective. Success with daratumumab has been the basis for expanded blood
disorder trials.
Since the red cell membrane has CD38 receptors, daratumumab often results in a positive indirect anti-human globulin test result. Daratumumab (anti-CD38 mediated) positive indirect anti-human globulin test can persist for up to six months following the last treatment. Prior to daratumumab administration, the drug manufacturer recommends a blood type and antibody screen.13 However, if blood is needed in the course of treatment, the blood bank is often challenged to determine if there is a new underlying antibody. Dithiothreitol (DTT) is limited and may not provide a clear understanding of any potential alloimmunization to antigens sensitive to enzyme treatment. Neutralizing substances to anti-CD38 is a potential tool in serological workups, but may not be available. But BGG can be the most beneficial, providing clarity of the patient’s genotype without laborious and expensive techniques. BGG directed at the patient’s DNA is not susceptible to interference by transfused products or infused drugs.
Changing the paradigm from phenotype to genotype determinants
In the last five years, the Food and Drug Administration (FDA) has approved two red cell genotyping assays based on detection of single nucleotide polymorphisms (SNPs) on selected genes. In 2014, the pre-market approval of the Immucor PreciseType HEA Molecular BeadChip Test was cleared. This device is indicated for the molecular determination of allelic variants that predict 36 erythrocyte antigen phenotypes.
In 2018 the FDA approved ID CORE XT, a molecular-based assay used in blood transfusion medicine to help determine blood compatibility. The ID CORE XT has its origin with the BloodGen project, funded by the European Commission (2003-2006) to investigate DNA array-based methodology for genotyping blood donors or patients. The FDA approved assay can be used to determine blood donor and patient non-ABO red blood cell (RBC) genotypes. The FDA identified the ID CORE XT as the only blood group genotype assay cleared to genotype the polymorphisms and predict the allele genotypes and antigen phenotypes of the blood group systems listed in Table 2 (available online), as an alternative to serology. In December 2018, the FDA issued their final guidance on, “Labeling of Red Cell Units with Historical Typing Results.”14
Table 2 provides a comparison of the two FDA approved HEA molecular assays. Package inserts and FDA’s Summary of Safety and Effectiveness are available online at www.fda.gov. The key differences are:
HEA BeadChip Kit (PreciseType) is an in vitro diagnostic test intended for the molecular determination of allelic variants that predict erythrocyte antigen phenotypes predictive phenotypes as final result. The ID CORE XT is FDA approved for reporting genotypes. Predictive phenotypes do not require additional serology for labeling or reporting.
- ID CORE XT predicts the Miltenberger (Mia, MNS7) antigen. Detection of the Miltenberger (Mia, MNS7) antigen is a discrete antigen of the MNS system. The Mia is the product of a hybrid gene, probably a mutation or crossover events of glycophorin A and glycophorin B responsible for the unique glycophorin hybrid peptide, Gp.Mur phenotype. In the Japanese and white population, the frequency is very low, 0.006 percent and 0.0098 percent, respectively. The frequency is much higher in other parts of Asia, such as Chinese (seven percent), Thai (10 percent) and even higher in indigenous tribes in Taiwan (∼21.2-88.4 percent). Antibodies to the Mur and the Mia antigen can cause hemolytic transfusion reactions (HTR) and HDFN.15
- PreciseType detects the Hemoglobin S mutation in the Beta Globin gene. Results from this mutation detection are not intended for diagnosis of SCD.
- PreciseType is predictive for Landsteiner and Weiner (LW) and Scianna antigens. However, antibodies to LW are not considered clinically significant and have not been implicated in HTR or HDFN. Antibodies to Scianna (anti-Sc1 and anti-Sc2) are not thought to be implicated in HTR or HDFN, but mild cases of HDFN have been reported. Antibodies to Scianna are detected by anti-human globulin technique and can be resistant to enzyme treatment.
- The use of molecular testing has evolved transfusion medicine. This is especially true regarding transfusion support for blood disorders dependent on transfusion therapy. The ability to go beyond serology and predict the genotype has helped to resolve serologic issues and focus on the exons responsible for the antigen structure. As molecular testing continues to improve—and the reality of Next Generation Sequencing becomes affordable—it is possible in the future that matching transfusion recipients and selected genotypically matched donors will be standard practice.
- Cellular determinants are controlled by nucleic acid Sequences
- Exons are part of the gene controlling coding amino acids for protein structures imparting for unique determinants
- Phenotype is the observed or biochemical characteristic resulting from the genotype and the environment
- Genotype is a description of the gene(s) in the DNA responsible for specific traits or characteristics
- Understanding molecular determinants has helped advance hematologic and oncologic care and treatment
- FDA has cleared assays for determination of genotype the polymorphisms and predict the allelic variants genotypes and erythrocyte antigen phenotypes of blood group systems
REFERENCES
- Anstee DJ. The relationshop between blood groups and disease. Blood 2010 115:4635-4643.
- Storry JR, Olsson ML. The ABO blood group system revisited: A review and update. 2009;25(2):48-59.
- Lai L, Lee T-H, Tobler L, et al. Relative distribution of West Nile virus RNA in blood compartments: implications for blood donor nucleic acid amplification technology screening. Transfusion. 2012; 52:447-454.
- Williamson PC, Linnen JM, Kessler DA, et al. First cases of Zika virus–infected US blood donors. Transfusion. 2017; 57:770-778.
- Da Cunha Gomes EG, Machado LAF, de Oliveira LC, Neto JFN. The erythrocyte alloimmunisation in patients with sickle cell anaemia: a systematic review. Transfusion Medicine. 2018; doi: 10.1111/tme.12543.
- Gehrie EA, Ness PM, Block EM, Kacher S, Tobian AAR. Medical and economic implications of strategies to prevent alloimmunization in sickle cell disease. 2017; 57(9): 2267–2276.
- Kauf Tl, Coates TD, Huazhi L, Mody-Patel N, Hartzema AG. The cost of health care for children and adults with sickle cell disease. Am J Hematol. 2009; 84(6):323-327.
- Campbell-Lee SA, Gvozdjan K, Choi KM, et al. Red blood cell alloimmunization in sickle cell disease:assessment of transfusion protocols during two time periods. Transfusion. 2018;58;1588–1596.
- Schonewille H, Honohan A, van der Watering LM, et al. Incidence of alloantibody formation after ABO-D or extended matched red blood cell transfusions: a randomized trial (MATCH Study). Transfusion. 2016;56;311–320.
- Floch A, Tournamille C, Chami B, Pirenne F. Genotyping in Sickle Cell Disease Patients: The French Strategy. Transfus Med Hemother. 2018;45:264–270.
- Lane WJ, Westhoff CM, Gleadall NS, Aguad M, Smeland-Wagman R, Vege S, et. al. Automated typing of red blood cell and platelet antigens: Automated typing of red blood cell and platelet antigens. Lancet Haematol 2018; 5: e241–251.
- SG, Flegel WA, Westhoff CM, Denomme GA, Delaney M, Keller MA, et al. It’s time to phase in RHD genotyping for patients with a serologic weak D pheontype. Transfusion. 2015 Mar;55(3):680-689.
- Janssen Pharmaceutical Company of Johnson & Johnson. DARZALEX (daratumumab) Aproved by U.S. FDA: First Human Anti-CD38 Monoclonal Antibody Available for the Treatment of Multiple Myeloma. https://www.jnj.com/media-center/press-releases/darzalex-daratumumab-approved-by-us-fda-first-human-anti-cd38-monoclonal-antibody-available-for-the-treatment-of-multiple-myeloma. Accessed on December 20, 2018.
- Food and Drug Administration. Labeling of Red Blood Cell Units with Historical Antigen Typing Results; Guidance for Industry. 2018. https://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Blood/UCM534978.pdf?utm_campaign=What%27sNew2018-12-20&utm_medium=email&utm_source=Eloqua. Accessed on December 21, 2018.
- Hsu K, Chi N, Gucek M, Van Eyk JE, Cole RN, Lin M, Miltenberger blood group antigen type III (Mi.III) enhances the expression of band 3. Blood. 2009;114:1919-1928.
About the Author