Variations in crossing thresholds during real-time PCR when using auto-gain settings determined from the fluorescent signal in tube one
The ability to PCR multiple targets with a single master mix but different primers and probes under identical cycling conditions with a single real-time PCR instrument allows for an efficient and streamlined workflow. We validated a variety of sample types for multiple viral targets as single assays that could then be run individually or concurrently.
Assay design
Using RealStar Analyte Specific primers and probes and RealStar QAM positive controls for herpes simplex virus (HSV)1, HSV2, varicella zoster virus (VZV), adenovirus, and parvovirus B19 (Altona Diagnostics), viral targets were validated individually by real-time PCR on the Rotor-Gene Q MDx instrument (QIAGEN).
Samples were spiked with Altona internal control (IC) DNA prior to extraction on automated extraction platforms. Extraction instruments and kits used varied with sample types. Each PCR reaction contained 10µl DNA, 17µl GPR PCR Master 1.0, 1µl ASR target-specific primer, 1µl ASR target-specific probe, and 1µl water, except for HSV1/2, which contained 1µl each of HSV1 primer, HSV2 primer, HSV1 probe, and HSV2 probe. Cycling conditions for all targets were 95°C, 10 minutes followed by 45 cycles of 95°C, 15 seconds and 58°C, 60 seconds. Auto-gain optimization was used. All viral targets were detected in the green (FAM) channel except for HSV2, which was detected in the red (Cy5) channel. IC was detected in the yellow (JOE) channel.
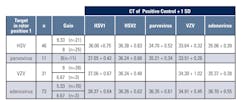
Upon implementation for clinical use, depending on what orders were received, HSV1/2, VZV, adenovirus, and/or parvovirus B19 were run concurrently. Crossing threshold (CT) reference ranges for positive controls were established for each viral target at a fixed threshold during validation of individual targets.
Observations
CT shifts of 1-2 cycles for positive controls were observed when more than one viral target was run because initial reference ranges for positive controls were established by running viral targets individually. CT shifts were due to differences in auto-gain settings which are determined from the fluorescent signal in tube one in the Rotor-Gene instrument and are set for the green channel.1 “Gain” values are displayed on Rotor-Gene reports and are low if the fluorescence signal is strong and high if the fluorescence signal is weak.
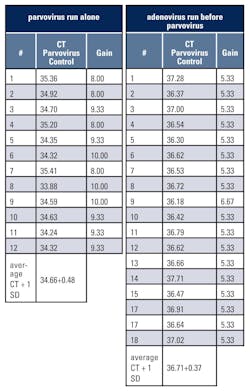
Table 1 shows CT and gain values of the parvovirus positive control when parvovirus was run alone (12 successive runs) or when adenovirus was run before parvovirus (18 successive runs). The average CT value + 1 standard deviation (SD) of the parvovirus positive control increased from 34.66+0.48 to 36.71+0.37 when adenovirus was run first. Gains were 8, 9.33, or 10 when parvovirus was run alone and typically 5.33 when adenovirus was run before parvovirus.Conclusion
If the auto-gain setting is used on the Rotor-Gene and multiple targets are run concurrently, the target with the lowest gain identified from individual runs should be in rotor position 1. Positive control CT ranges should be established from both individual runs and runs where the target with the lowest gain is run in position 1. It should be noted that the CT shifts observed did not affect patient results and that these recommendations may vary with different real-time PCR instruments.
REFERENCE
The authors wish to thank Drs. Karin Rottengatter and Konstance Knox for their assistance and helpful advice.