Automation advances microbiology to operational excellence
CONTINUING EDUCATION
To earn CEUs, visit www.mlo-online.com under the CE Tests tab.
LEARNING OBJECTIVES
1. Describe the trends in healthcare which have led to the changes that the clinical microbiology laboratory is facing.
2. List and describe the technological advances that have contributed to the evolution of the clinical microbiology laboratory.
3. Discuss the factors that must be addressed when implementing automation into the clinical microbiology laboratory.
4. Describe new and upcoming advancements that will continue to evolve the clinical microbiology laboratory.
The past few years have seen significant changes in clinical microbiology. On the regulatory front, as more assays receive FDA clearance, labs increasingly have the option to replace laboratory-developed tests (LDTs) with commercially available IVD tests. Respiratory panels are a good example. Second, technological advances continue to reduce turnaround time drastically, and, at the same time, a better understanding of how test results can be used to improve care is raising physician expectations. For example, while a turnaround time of eight to 24 hours for a flu test was previously acceptable, a 20-minute to two-hour time to result is now standard of care. Likewise, while a turnaround time of >24 hours for generating a C. difficile result was previously acceptable, a 1½- to three-hour time to result is becoming the standard of care.
Patient management, the decision to admit, and infection control measures often hinge on test results, with significant health outcome and cost ramifications. Microbiology results, once viewed as confirmatory and often delivered after patient management decisions were made, are now integral to the clinical workflow. Patient-centric care, the mantra of today’s healthcare, translates into patient-centric testing in the microbiology lab, with far-reaching implications for the lab workflow, how the lab is structured, and even how patient specimens are delivered to the lab.
Along with this trend toward patient-centric testing has come a massive shift toward automation, driven by the need to better support clinical decisions with high-quality, timely results efficiently and cost-effectively. This trend is enabled by advances such as molecular diagnostics, digital microbiology, and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS).1 These advances open the door to greater standardization of processes and results, automation, and a new level of operational excellence and performance. Efficiency gains in the lab are especially needed, with continuing pressure on reimbursement due to the Protecting Access to Medicare Act of 2014 (PAMA).
Enabling technologies continue to evolve
Clinical microbiology has traditionally been associated with a diversity of patient specimens and transport media, lengthy culture, and visual results requiring human review. These factors, all of which hamper standardization, a prerequisite of automation, have left the microbiology lab in the dark ages. All this is changing with recent technological advances that are enabling automation at a time when microbiology most needs it—to meet the multiple challenges of today’s healthcare environment, declining reimbursement, and the shortage of trained personnel.
Molecular diagnostics has truly been a transformational force in microbiology. The sensitivity it offers has allowed us to detect and identify more organisms faster, without having to wait for culture results. Mycoplasma is a case in point. Another example is testing for sexually transmitted infections like Chlamydia trachomatis and Neisseria gonorrhoeae. In many ways, molecular diagnostics has made diagnostics more relevant. Previously, a patient suspected of chlamydia or gonorrhea would be treated empirically. Today, with faster time to result, treatment can begin after the pathogen is properly identified, reducing the unnecessary use of antibiotics. A study comparing clinical and economic outcomes before and after initiation of molecular testing of methicillin-resistant Staphylococcus aureus (MRSA) in positive blood cultures demonstrated a mean reduction of 6.2 days in hospital stays and a $21,387 savings when molecular testing was combined with timely infectious disease consult and appropriate antibiotics use.2
Molecular diagnostics offers other advantages—the availability of automated platforms, options in standalone specimen preparation and handling systems, and connectivity to the chemistry lab. All of these can help advance clinical microbiology through automation and better integration with the rest of the healthcare system.
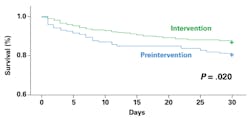
A second transformational technology is MALDI-TOF, first commercially available for clinical microbiology in 2009. MALDI-TOF provides a cost-effective, standardized, and rapid method for identification of clinically significant bacteria, fungi, and mycobacteria. Importantly, MALDI-TOF procedures are relatively simple and do not vary based on organism. The integration of MALDI-TOF into the clinical microbiology workflow has decreased time to organism identification from days to hours. The impact on patient care is significant. One study on adult patients with bacteremia and candidemia demonstrated that the combination of MALDI-TOF diagnostic testing and an antimicrobial stewardship team decreased time to effective and optimal antibiotic therapy, which translated into lower mortality rate, shorter intensive care unit stays, and reduced recurrent bacteremia (Figure 1).3 For the first time, standardizing the identification process of bacterial isolates is now possible, enabling automation, improving workflow, and increasing efficiency.
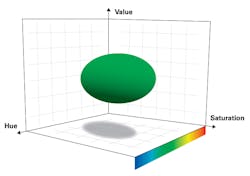
The evolution of digital microbiology is changing the way in which culture plates are read, impacting not only turnaround time but also quality of results.4 Digital microbiology integrates automated imaging systems to capture images of culture plates at specified time intervals, robotics to move specimens across the lab, and software to process captured images and determine the presence of positive colonies for review and confirmation. For example, with digital imaging software, preset color thresholds, as defined by hue, saturation, and value, are employed to detect growth in chromogenic agar (Figure 2). In this way, negative plates can be identified for quick screening to confirm negative results, while only positive plates are reviewed one by one. This reduces substantially the number of plates reviewed and ensures better use of laboratorian time when trained personnel are scarce. The ability to rule out negative plates quickly not only saves personnel time but can expedite clinical decisions and reduce unnecessary intervention.
The advent of liquid transport media has been highly influential in improving specimen quality and enabling automation. Elution of specimens from newer flocked-style swabs into liquid phase has been shown to increase the release of viable organisms from the swab, improving sensitivity. And the liquid-based transport enables inoculation of the specimen with automated liquid-based specimen processors, an important step toward greater efficiency and standardization.5
Microbiology automation—indications of success
While microbiology automation is only at the early stages of implementation—an estimated 10 percent of labs in the U.S. having adopted some automation technology—quantifiable improvements in turnaround time, productivity, and quality have been reported.
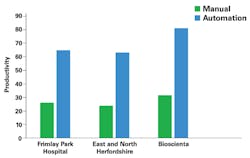
In a multi-center study, molecular diagnostics was shown to reduce time to results for MRSA testing from 24 to 48 hours to less than two hours.6 A study comparing routine microbiology with automated identification and susceptibility testing on patients with positive blood cultures found a mean reduction of 13 hours in turnaround time for identification and 20 hours for susceptibility results.7 The combination of automation of front-end specimen processing with MALDI-TOF yielded a 30.6-hour time gain per isolate compared with conventional microbiology.8 Another study reported that productivity, as measured by number of samples processed per FTE per day, showed a 2.5- to 2.7-fold improvement in three hospitals after the automation of plate handling and incubation (Figure 3).9
Automation enables workflow optimization, removing unnecessary delays and better utilizing the skills of trained lab professionals. One example is the use of an automated system for plating and streaking of urines and other liquid specimens, while trained personnel perform Gram stain review and the processing of more complex specimens. This saves time as well as improves consistency in urine plating while decreasing monotonous repetitive tasks for laboratorians. Another example is the ability to read cultures using an automated imager following specified incubation time rather than wait for the availability of personnel, a change that not only shortens time to result but also improves consistency of results and ensures that plates are not incubated beyond recommended duration, which can compromise specificity.10,11
Quality improvement is an important benefit of automation, which allows standardization of techniques and reduction of human errors. A recent study demonstrated that automating specimen processing, specimen incubation, imaging of MRSA growth, and automated analysis reduced the number of false negatives. Compared with manual screening, sensitivity was 100 percent across three sites using three different commercially available culture media, and specificity ranged from 90.7 percent to 92.4 percent.4 Automated analysis identified all positives identified manually, while 2.9 percent of the discrepant results (identified as positive by automated analysis and negative by manual reading) were found to be positive upon manual re-examination. In another study comparing automated imaging and analysis with manual review of 92 urine specimens, automated analysis identified four additional positive cultures missed by manual reading due to overlooking a pathogen in heavily mixed cultures.12
Related to quality improvement are traceability and documentation. By automating the confirmation of multiple patient identifiers at multiple steps from specimen transport vessel to plates, the lab can prevent medical errors due to incorrect patient identification. In combination with inexpensive electronic data storage, digital microbiology facilitates documentation with image archiving for further review, for correlating Gram stain images with cultures, for use in teaching, and to share with inquiring physicians.
Perhaps the most important benefit of automation is its value in optimizing the lab workflow, which in turn positively impacts the clinical workflow. For example, changing from reading plates when personnel can get to them to reading them when they are ready to be read can make all the difference in optimizing turnaround time. In turn, this means availability of results when physicians need them. Knowing that the majority of specimens arrive at the lab between 8 PM and 2 AM, for example, the lab can plan to read plates until 10 PM, transition the staff to processing to get the plates to the incubator earlier, and start reading again at about 2 AM, when the plates are ready to be read by the automated imager. In this way, results from specimens that arrive the night before will be in the physician’s in-box the next morning. Another example is group A strep, with most specimens coming from Outpatient and arriving between 8 PM and 2 AM. With automated molecular diagnostics, results can be waiting for physicians the next morning. With the availability of technologies that offer group A strep results in 20 minutes, physicians have the option to diagnose and begin treatment while the patient is still in the office. These changes have a very positive impact on physician satisfaction ratings.
Implementing microbiology automation
Microbiology automation is not without challenges. Cost is a significant barrier to automation, with the initial investment in capital acquisition, facility modifications, and, importantly, IT and connectivity to integrate the entire process from test requisition and specimen collection to final result. Overcoming the initial resistance of lab personnel is also sometimes a factor.
While many labs have implemented some automation solutions, especially in molecular diagnostics, the majority of labs in the U.S. are still in the planning process or looking to build on what they have started. As with any significant initiative, thorough need assessment, planning, and securing the buy-in of all stakeholders are key. In this context, it is difficult to overemphasize the importance of two areas of focus in the planning stage—workflow and IT.
Automation cannot be done in a vacuum, and it is important to consider the overall workflow of the lab, not just specific processes being automated. Studies have shown that implementing automation without considering the overall workflow makes little impact on quality of care. To understand the overall lab workflow and the clinical workflow and how best to derive the maximum value from automation, labs need to review all facets of current lab operations. These include, for example, the mix of specimen types (i.e., whether processing can be automated), time they arrive at the lab, and expected time to result. A detailed snapshot of current hourly workload and staffing needs will guide how staffing might be redistributed after automation. The goal is to make sure there is clear understanding of how automation will be incorporated and how it will impact the overall workflow and, ultimately, patient care.
A second planning parameter that is sometimes left as an afterthought is IT. In the current imperfect world of connectivity, making sure that computer systems—from automation technology to MALDI-TOF to LIS to HIS—talk to each other is paramount. The need for integrated information flow cannot be overemphasized. Thus, getting the IT team involved from the beginning is a critical success factor. As higher-quality information is generated in a more timely manner, it is just as important to get it into the physician’s hands without delay.
Of course, there are myriad other factors to consider in planning automation—space planning is one example. Mapping out the post-automation workflow to reallocate staff time is another. A clear picture of the expected results and metrics for post-automation assessment will help track progress and guide adjustments once implementation begins.
Looking ahead
Even as labs are implementing the many advances that are reshaping clinical microbiology today, new technologies are on the horizon. One example is the quick identification of multidrug-resistant organisms and antibiotic susceptibility assessment directly from clinical samples, without the need for traditional enrichment, culture, or sample preparation processes. Currently, a new system is being evaluated in multiple sites across the U.S. The system has automation capabilities such as random access, familiar to the chemistry lab.
REFERENCES
- Ledeboer NA. Automation as a benefit in clinical microbiology. J Clin Microbiol. doi: 10.1128/JCM.00686-14.
- Bauer KA, West JE, Balada-Llasat J-M, Pancholi P, Stevenson KB, Goff DA. An antimicrobial stewardship program’s impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis. 2010;51:1074-1080. doi: 10.1086/656623.
- Huang AM, Newton D, Kunapuli A, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013;57:1237-1245. doi: 10.1093/cid/cit498.
- Faron ML, Buchan BW, Vismara C, et al. Automated scoring of chromogenic media for detection of methicillin-resistant Staphylococcus aureus by use of WASPLab image analysis software. J Clin Microbiol. 2016;54:620-624. doi:10.1128/JCM.02778-15.
- Bourbeau PP, Ledeboer NA. Automation in clinical microbiology. J Clin Microbiol. 2013;51:1658-1665.
- Peterson LR, Liesenfeld, O, Woods CW, et al. Multicenter evaluation of the LIghtCycler Methicillin-Resistant Staphylococcus aureus (MRSA) Advanced Test as a rapid method for detection MRSA in nasal surveillance swabs. J Clin Microbiol. 2010;48:1661-1666.
- Kerremans JJ, Verboom P, Stijnen T, et al. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J Antimicrob Chemother. 2008;61:428-435. pmid:18156278.
- Mutters NT, Hodiamont CJ, de Jong MD, Overmeijer HPJ, van de Boogard M, Visser CE. Performance of Kiestra Total Laboratory Automation combined with MS in clinical microbiology practice. Ann Lab Med. 2014;34:111-117, http://dx.doi.org/10.3343/alm.2014.34.2.111.
- Matthews S, Deutekom J. The future of diagnostic bacteriology. Clin Microbiol Infect. 2011;17:651-654. doi: 10.1111/j.1469-0691.2011.03512.x.
- Joubrel C, Gendron N, Dmytruk N, et al. Comparative evaluation of 5 different selective media for Group B Streptococcus screening in pregnant women. Diagn Microbiol Infect Dis. 2014;80:282-284. doi: 10.1016/j.diagmicrobio.2014.08.005.
- Perez JM, Cavalli P, Roure C, Renac R, Gille Y, Freydiere AM. Comparison of four chromogenic media and Hektoen agar for detection and presumptive identification of Salmonella strains in human stools. J Clin Microbiol. 2003;41:1130-1134. pmid:12624041.
- Riebe KM, Buchan BW, Ledeboer NA. Validation of the WASPLab system for urine specimens. Poster presented at American Society for Microbiology 115th General Meeting, 2015.