How NGS panel advancements help us find and fight colorectal cancer
Colorectal carcinoma (CRC) is not only the fifth most deadly cancer but also one of the most commonly diagnosed cancers worldwide.1 Therefore, it is imperative that we develop better methods to diagnose and treat CRC. Next generation sequencing (NGS) technologies have been advancing the use of genomic sequencing for clinical applications, including the simultaneous study of mutations in high-penetrance colorectal cancer genes.
NGS panels — also referred to as targeted gene sequencing panels — offer practical diagnostic tools that help inform personalized treatment. For example, while monoclonal antibodies (such as cetuximab and panitumumab) can block tumor progression by targeting the epidermal growth factor receptor (EGFR), there are some tumors with specific mutations that do not respond to these therapies.2 By focusing on the select set of genes or gene regions carrying these mutations, NGS panels offer a rapid way to identify whether a patient will respond to treatment.
Although NGS panels are now in routine use for a wide range of research and clinical applications, there remains significant hurdles — including how to handle challenging formalin-fixed paraffin-embedded (FFPE) samples, GC-rich target regions, and tandem repeats. However, recent advances in workflow design are enhancing the coverage, uniformity, accuracy, and sensitivity of colorectal cancer NGS panels.
Dealing with difficult samples
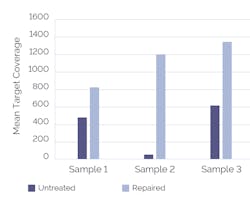
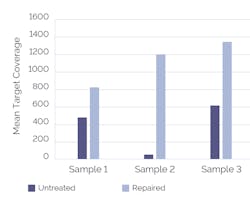
The use of an upstream FFPE repair step — which can remove a broad-range of damage including nicks, gaps, oxidized bases, and cytosine to uracil deamination — has been shown to significantly improve sequencing performance (Figure 1). Additionally, the choice of target enrichment strategy can impact how well an assay performs.
Which enrichment strategy?
Target enrichment is one of the most important pre-sequencing steps of NGS colorectal cancer panels. The two most used target enrichment strategies are PCR-based (amplicon) and hybridization-based assays. Selecting the right target enrichment strategy is critical, since it will influence the outcome of sequencing results — and ultimately, research conclusions and clinical decisions. For example, PCR-based enrichment is known to be particularly susceptible to contaminants found in FFPE materials, while hybridization-based assays are typically more robust.
Challenging target regions
Researchers often find themselves trying to work with difficult sequences. For example, TP53 has been identified as a mutated cancer gene, including in CRC. However, sequencing is frequently confounded by the GC-rich content of the exons (5-8) where TP53 point mutations typically occur. Moreover, non-uniform, incomplete, and non-specific amplification caused by varied GC content is a common problem associated with PCR-based target enrichment. Careful design of hybridization baits has led to efficient capture, resulting in much more uniform coverage than amplicon assays (Figure 2). The ability to use longer probes also lends itself to tolerance of sequence variation, such that all alleles can be captured equally.
Other challenging target regions include those containing internal tandem repeats. However, by designing additional hybridization probes — both upstream and downstream of the repetitive region — sequences around the repeat areas can be captured. Additionally, the potential use of longer probes in hybridization-based enrichment assays can read through short (up to 100 bp) repetitive regions.
Getting rid of the noise
As NGS panels transition into routine clinical diagnostics, reducing false negatives and false positives — the ‘noise’ — is critical. For example, if a patient is given a false negative/positive result, they may be considered for a treatment that will ultimately be ineffective.
While poor coverage of the target site represents the primary cause of false negatives, improving uniformity of enrichment by well-designed hybridization assays can reduce their incidence. On the other hand, the most common cause of false positives are artefacts introduced during PCR. Since artefacts will be amplified exponentially in line with increasing PCR cycles, the much lower number of cycles required for hybridization assays are favorable.
These minimal PCR cycles needed for hybridization assays translate as reduced ‘noise’ and, in turn, higher detection sensitivity of low frequency variants. Indeed, one study compared commercially available kits for the detection of 24 mutations associated with cancer, and the hybridization-based assay showed the highest sensitivity, able to detect all known variants (100% sensitivity) as well as two novel mutations.3 The more comprehensive profiling for all variant types, both novel and known, lends itself to both research and clinical applications.
It is time for PCR to shine – or is it?
One of the major benefits of PCR-based amplification is its more streamlined library preparation, compared to hybridization-based strategies. However, changes in hybridization-based workflow design, such as combining the end-repair and adaptor ligation steps, mean that it is now possible to load more samples onto a sequencer within a single day.
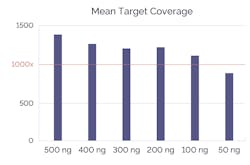
PCR-based amplification typically requires significantly less DNA input than hybridization assays. However, recent design advances to hybridization mean that some now need considerably less starting material (Figure 3). Moreover, while smaller starting concentrations might sound desirable, it is important to consider the effect on sequencing results.
Small amounts of input DNA mean less template DNA is available, and less template DNA can lead to high duplication rates. With hybridization assays, duplicates can be removed swiftly during the bioinformatics process. This is not a possibility with amplicon data without the use of molecular barcodes, resulting in skewed data due to the over-amplification of a small number of fragments.
The outlook - from research to clinic
Multiplex NGS panels are set to change diagnostics, prognosis, and treatment options for colorectal cancer patients, affording the ability to effectively screen patients for numerous mutations simultaneously. Moreover, as we see a rapid improvement in NGS technology, it is likely that there will be a significant reduction in sequencing cost and the ability to test specimens from many patients on the same assay, making the jump from research lab to clinical lab much more manageable.
Importantly, we must also consider how changes in workflow design can significantly affect sequencing outcomes, particularly during target enrichment. While choice of target-enrichment strategy is fundamentally dependent on goals and resources, hybridization-based assays offer more confidence in calling all variants, with less noise. This will ultimately impact both research and clinical outcomes.
References
- Sung H, et al. Global cancer statistics 2020: GLOBOCAN Estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021 May;71(3):209-249. doi: 10.3322/caac.21660.
- Zhao B, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2017 Jan 17;8(3):3980-4000. doi: 10.18632/oncotarget.14012.
- Burghel GJ, et al. Towards a next-generation sequencing diagnostic service for tumour genotyping: A comparison of panels and platforms. Biomed Res. Int. 2015;2015:478017.
About the Author

John Townes, BSc
is Strategic Product Manager for SureSeq targeted NGS products at OGT. Townes earned his BSc (Hons) in Biological Sciences from The University of Reading, UK.