Molecular assay design strategies for impactful patient management
Laboratory medicine has made significant advances over the last two decades. Viral load assays have evolved from nested PCR assays to transcription-mediated amplification to real-time PCR and continue to evolve with methods like digital PCR. This advancement has resulted in more sensitive viral load assays with a broader dynamic range, which allows physicians a better understanding of a patient’s response to therapy and disease progression.
One example of improved patient management with the advancement of molecular diagnostics has been the management of HIV-1 patients. According to the current treatment guidelines, treatment success is defined as HIV-1 RNA concentration below 75 copies/mL.1 The definition of treatment failure varies by global, national and country-specific guidance, but is usually defined as HIV-1 RNA concentration between 50–1000 copies/mL.1-3
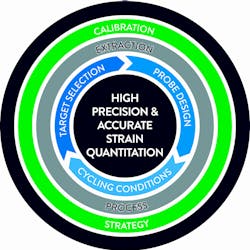
These treatment cut-offs are at the low end of the dynamic range of the real-time PCR assays where precision and reproducibility may be challenged. Factors which could influence viral load assay performance are automated platform, nucleic acid extraction chemistry, PCR primer/probe selection and design, PCR amplification conditions and assay calibration strategy. Here we will review different design approaches and their potential impact on assay performance and clinical outcome. (Figure 1)
Extraction process
One should carefully consider sample type and analyte when selecting extraction chemistry. For example, in cases of HIV-1, the Department of Health and Human Services (DHHS) advises that HIV-1 RNA should be used as a marker of HIV-1 viremia.1
The distinction between HIV-1 RNA and proviral DNA is important as the latter is not a marker of active viral replication, but rather of latent reservoir release from the virus, which has already been integrated into the cells. An extraction chemistry that purifies both HIV-1 RNA and proviral DNA would not provide an accurate representation of true viral replication to the physician; therefore, in the instance of this virus, only the extraction chemistry that specifically purifies HIV-1 RNA should be used in order to exclude proviral DNA from downstream quantitation.
Alternatively, the use of total nucleic acid (TNA) extraction chemistry for quantitation of HIV-1 could potentially lead to false-positive results or inaccurate quantitation, which would have adverse effects on patient management. TNA extraction chemistry can be utilized for more difficult sample types such as urine or stool or for assays that need to detect both RNA/DNA targets.
Target selection
The extreme viral diversity encountered amongst HIV, HBV and HCV strains are of utmost concern for ensuring that molecular diagnostic (MDx) tests give the correct result regardless of the strain(s) present in a sample.
To address this concern, the optimal assay design should start with the selection of primer/probe target in a genetically conservative region where viral diversity will have the least potential impact. The genetically conservative regions can only be determined by comparing sequences from diverse viral strains.
To meet this need, a global surveillance program is critical to comprehensively assess the existing viral diversity in circulation for HIV, HBV and HCV strains.4 In a surveillance program, sequences generated from strains collected in geographically diverse regions are used to determine which regions of a viral genome are most well conserved and amenable to detection by a molecular diagnostic test. By using circulating viral sequences to inform assay design, the risk of strains being missed by a test is significantly reduced.
Probe design and PCR cycling conditions
In addition to the selection of the target region, probe design and PCR cycling conditions are critical as we look to ensure accurate viral load quantitation. Probe design needs to be tolerant of naturally occurring polymorphisms and in turn, potential mismatches that may occur within the given target region. Over time, probe technology has evolved, as has the PCR methodology. The wider range of MDx applications based on PCR technology beyond viral quantitation require utilization of different probe designs. Minor groove binding probes are typically used for genotyping and detection of single nucleotide polymorphism.
TaqMan probes are often used in assays where target regions have a high degree of conservation. Partially double-stranded probes, which are better at tolerating high levels of genetic heterogeneity mainly due to probe length and binding conditions, are utilized in areas where there is a high degree of heterogeneity.5
In addition to probe design, cycling conditions are oftentimes the subject of design optimization, in order to achieve the performance requirement (e.g. tolerance of potential mismatches.) Phase-matched cycling is one approach that incorporates cycles at a lower temperature to reduce initial stringency, which is followed by cycles at high temperatures to preserve specificity. This approach mitigates the adverse effect on sample quantitation by potential primer mismatches. In the case where it is difficult to avoid the significant impact of rare polymorphisms, the detection of a second target may be incorporated in the assay design. When sequence diversity affects the detection of one region of the viral genome, the detection of a second region ensures that an accurate result will be achieved. Given the complexity of molecular assays and high level of viral diversity, a comprehensive approach of utilizing global surveillance and target-specific probe design should be considered when designing molecular assays. Simply adapting one strategy such as dual-target may provide a false sense of security.
Calibration strategy
Calibration strategy is an integral component of molecular assay design and is critical to ensuring reproducibility across a wide dynamic range. Most quantitative methodologies for therapy management currently use an external calibration curve to determine the concentration of an analyte.
In these assays, the analyte signal in a patient sample is compared to a set of samples with a known concentration and a simple linear regression (y=mx+b) is used to calculate the viral load. This approach typically uses calibrators that are processed as patient specimens through the entire process, allowing for calibration of both extraction and amplification reagents and instruments.
Alternatively, calibrators that are not processed through the extraction could be used, however, this approach carries the risk that difference in recovery or a change in the reagent composition wouldn’t be accounted for, potentially leading to differences in quantitation.
Another less frequently used strategy is an internal quantitative standard. This application uses a 3rd order polynomial regression line (y = ax3 + bx2 + cx + d) across the linear range with an allowable maximum difference from linearity. Previous studies have suggested that the acceptable allowable difference from linearity for some of the assays was ± 0.2 Log10.6 This allowable difference from linearity and the calibration approach also explains the bias often observed between methodologies, as well as larger imprecision at the low end of the dynamic range where clinical decisions are often being made.
Conclusion
Accurate and precise molecular assay performance is critical for appropriate therapy management. Inaccurate viral load results could lead to inappropriate management and the patient may be left on failing therapy. This misdiagnosis has much broader implications than management of a single patient as it could also lead to an increase in transmission of the resistant viral strain. From a therapy management perspective, a virus with mutations associated with resistance is more challenging to treat with narrower therapy options. In addition, false-positive rates can add unnecessary cost for repeat testing or more expensive resistance testing, as well as anxiety for the patient and the physician.
REFERENCES
- Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV developed by Department of Health and Human Services. https://aidsinfo.nih.gov/guidelines). Accessed September 2019.
- European AIDS Clinical Society (EACS). EACS Guidelines Version 9.1 October 2018.
- National Department of Health, South Africa, April 2015, accessed August 2, 2017: https://aidsfree.usaid.gov/sites/default/files/tx_south-africa_pmtct_2015.pdf.
- Brennan CA, Bodelle P, Coffey R, et al. HIV global surveillance: foundation for retroviral discovery and assay development. Journal of medical virology. 2006;78 Suppl 1:S24-29.
- Luk KC, Devare SG, Hackett JR, Jr. Partially double-stranded linear DNA probes: novel design for sensitive detection of genetically polymorphic targets. Journal of Virological Methods. 2007;144(1-2):1-11.
- Vermehren J, Colucci G, Gohl P, et al. Development of a second version of the Cobas AmpliPrep/Cobas TaqMan hepatitis C virus quantitative test with improved genotype inclusivity. Journal of Clinical Microbiology. 2011;49(9):3309-3315.
Acknowledgment: Author would like to acknowledge and thank Mary Rodgers and Shihai Huang for their review of this article.
About the Author
Danijela Lucic, PhD
received her PhD in Biochemistry from Rush University Medical Center, Chicago and completed her postdoctoral training at University of Illinois. She joined Abbott Molecular in 2006 and is part of the Global Scientific Affairs team with responsibilities for HIV-1, Hepatitis, and Transplant products. During her tenure at Abbott she has been involved in translation research of molecular diagnostics and their impact on patient management.