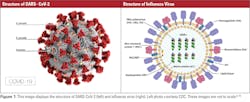
Comparing the structure, diagnosis, and prevention of SARS-CoV-2 and influenza
For a printable version of the September CE Story and test go HERE or to take test online go HERE. For more information, visit the Continuing Education tab.
LEARNING OBJECTIVES
Upon completion of this article, the reader will be able to:
1. Describe the structural differences between SARS-CoV-2 and influenza virus A or B (IAV or IBV).
2. Describe how antigenic drift and antigenic shift occur for IAV or IBV and SARS-CoV-2.
3. Describe the types of lab assays available for detecting an infection with SARS-CoV-2 or IAV and IBV, as well as tests used to monitor the health status of patients with COVID-19.
4. Describe the vaccines available to prevent disease from SARS-CoV-2 or IAV and IBV.
With COVID-19 being a respiratory disease that has similar symptoms to the influenza virus, and while the two viruses share some similarities, the differences are clinically significant. The severity of both viruses varies from person to person, with some people acting as asymptomatic carriers, and others receiving treatment in the intensive care unit on ventilators. The pandemic taught the scientific and medical communities more about COVID-19 than they knew in late 2019. Everything learned throughout the pandemic has led to the development of reliable laboratory testing and multiple vaccine options for COVID-19.
While COVID-19 is caused by SARS-CoV-2, influenza is a common seasonal illness caused by influenza virus A or B (IAV or IBV). The influenza virus was first isolated in the laboratory in 1932 but has caused pandemics and epidemics every year, which is similar to what we experience with seasonal influenza activity today.1 One of the most well-known outbreaks of the influenza virus was the 1918 Spanish flu, which was an outbreak of H1N1, or swine flu. The flu epidemic occurs every year in the United States in the fall and winter months. Like COVID-19, symptoms and severity of influenza vary from person to person. Getting a yearly flu vaccine helps reduce the risk of illness from the seasonal influenza virus. In 2017, the World Health Organization (WHO) reported that 650,000 people worldwide die due to flu-related respiratory illness each year. The data from the WHO includes low- to middle-income countries without the same access to preventative and acute medical care as is available in developed countries such as the United States.2
A comprehensive analysis of COVID-19 and influenza is necessary to differentiate between these two viruses regarding structure, identification, and vaccine development. Some of the most important similarities and differences are seen in the structure, symptoms, transmission, rate of mutation, laboratory testing, and available vaccinations.
Structure
Coronaviruses are characterized as positive single-stranded RNA enveloped viruses (ssRNA).3 The genome of positive sense ssRNA viruses can be used as messenger RNA (mRNA). This means that the genome does not have to be transcribed as mRNA first. SsRNA viruses are the least complex form of viruses and only contain information that is needed to infect a host cell. This type of virus replicates the nucleocapsid and the genetic material that will be contained in the nucleocapsid at the same time. Due to the ssRNA structure of coronaviruses, they rely greatly on host mechanisms for replication. Replication of the genome happens spontaneously, which does not require energy or adenosine triphosphate (ATP). This differs from DNA or RNA transcription where ATP is required for replication.4 Coronaviruses range in size from 26 to 32 kb, which makes them the largest single-stranded RNA virus. Glycoprotein projections on the surface of the virus give the virus a crown-like appearance that is visible using electron microscopy (EM).3 The basic structures of SARS-CoV-2 and influenza virus are shown in Figure 1.
On the other hand, the influenza virus is a negative ssRNA virus that has glycoproteins that project on the surface of the virion. Since the influenza virus is a negative ssRNA virus, the ssRNA is not transcribed directly because the genetic material is made of the complementary strand of RNA. This means that to replicate, the complementary RNA strand must be transcribed into the template strand first. Influenza A (IAV) and B (IBV) are 12kb in size, which is typical for negative ssRNA viruses. The genome of IAV is made up of eight different segments of negative ssRNA. The main proteins found in IAV are the hemagglutinin (HA) and neuraminidase (NA) proteins, which make up the projections on the surface of the virion. In the envelope of IAV, HA, NA, and the matrix-2 (M2) proteins are integral proteins on the membrane of the virion. An integral protein is essentially locked into the membrane of the virus. HA, NA, and M2 overlay a matrix of the matrix-1 (M1) protein, which encloses the virion core. IBV is similar to influenza A, but the NB and BM2 proteins replace M2. Both HA and NA are responsible for categorizing IAV into subtypes that contribute to the infectivity and mutation rate of the virus that will be discussed in later sections.8
One key difference between coronavirus and influenza is the ssRNA that contains the genetic material for each virus. Coronavirus has positive sense ssRNA; whereas, influenza has a negative sense ssRNA. The difference between these two types of genetic material is that the positive sense genome can act as mRNA and be directly translated into proteins by the ribosomes of the virus. On the other hand, negative sense ssRNA in the influenza virus does not have this ability.3, 8 To produce mRNA, the negative sense ssRNA relies on RNA-dependent RNA polymerase to transcribe the RNA strand that codes for mRNA and to make the corresponding proteins.8 Another difference between COVID-19 and IAV is genome complexity. As mentioned earlier, coronaviruses are the least complex viruses; whereas, IAV has a more complex genome. Genomic size is also a difference between each virus because coronavirus is 32kb; whereas, the influenza virus is 12kb.3, 8
Rate of viral mutations
Influenza is well known for its predisposition to antigenic drift and antigenic shift. Antigenic drift is the result of point mutations in the amino acid sequence that makes up the antigens HA and NA, which are presented on the viral lipid envelope. Mutations can result in the expression of different HA or NA glycoproteins. Antigenic shift occurs in the virus when there is an abrupt change to the glycoproteins on the surface of IAV. This can occur when IAV infects a species of animal that is not the intended host. H1N1, or “swine flu,” is an example of this because the virus was passed from swine to humans. In this scenario, pigs are the intended host, and humans are the accidental host. The H1N1 outbreak was the cause of concern, as the surface antigens HA and NA were different from previous antigens. Due to this, humans that became infected with the H1N1 variant of IAV did not have immunity because their immune systems did not have antibodies for the respective antigens on H1N1.11
COVID-19 can also undergo antigenic drift and antigenic shift over time. Two examples of prominent variants are SARS-CoV-2: B.1.1.7, which began in the United Kingdom (UK), and B.1.351, which was discovered in South Africa. Variant B.1.1.7 has a mutation in the receptor-binding domain, which binds to the angiotensin-converting enzyme 2 (ACE2) receptor of the host. The mutation in this variant is at position 501 of the spike protein, where asparagine is replaced with tyrosine. This variant has also shown an increase in transmissibility compared to the parent strain.12 Due to the increase of transmissibility of the B.1.1.7 variant, the virus strain was the most prominent strain in the United Kingdom at one point. The B.1.351 variant has multiple spike protein mutations. Both the B.1.1.7 and B.1.351 variant are more transmissible than the parent strain of COVID-19. The now-dominant Delta variant, or B.1.617.2, which first emerged in India, also is more transmissible.13
One mechanism that contributes to the high mutation rate of RNA viruses is RNA polymerase. RNA polymerase is an enzyme responsible for proofreading the RNA of a genome, before the RNA gets translated for replication. Since RNA viruses IAV and COVID-19 lack an effective proofreading functionality to ensure that the genome is being replicated accurately, they lend themselves to high mutation rates and the formation of different strains or variants. In IAV, the mutation rate ranges from 1.5^-3 to 7.2^-5/ bp.14 This means that, on average, for every 667 to 13,888 base pairs translated, there could be a mutation. While the errors from RNA polymerase contribute to the creation of new variants of existing viruses, RNA polymerase may be a detriment to the survivability of the virus. 14
Laboratory testing and diagnosis
Infection of COVID-19 or IAV may be first recognized by characteristic symptoms, but the only way to confirm the suspicion of infection is through laboratory testing, combined with a patient’s presentation of symptoms. There are two common methods of testing for COVID-19 and IAV: polymerase chain reaction (PCR) and antigen testing. There is also serological testing available to determine if a patient recently had an infection, which is determined by detecting antibodies.
Antigen testing identifies antigens from the virus that are in the host, which can be used to determine if there was a previous infection of COVID-19 or influenza. While molecular testing is used to confirm a COVID-19 diagnosis, other laboratory tests help providers manage patients with severe COVID-19 infections. According to an article published by Justin Jones, PhD, in Medical Laboratory Observer, COVID-19 patients show elevated levels of procalcitonin (PCT), Interleukin-6 (IL-6), D-dimer, ferritin, cardiac troponin, and N- terminal pro-B-type natriuretic peptide (NT-proBNP), compared to healthy individuals. 15
Molecular testing
PCR is a nucleic acid amplification process that detects the presence of a target genetic sequence by making millions of copies of the desired target in a matter of hours in vitro. This methodology uses the same components as in vivo replication, which includes DNA polymerase and a primer sequence. For viral nucleotide sequence detection, reverse transcriptase (RT) is also present to transcribe the RNA into complementary DNA (cDNA) for detection of the target sequence. In vitro RT-PCR replication differs from in vivo replication because in vitro RT-PCR targets a specific nucleotide sequence from the template strand of DNA, rather than duplicating the entire DNA strand.16 If the target sequence is present in the sample, the reagents used in the testing will fluoresce, indicating a positive result in the RT-PCR method of testing.
According to an article published by the College of American Pathologists (CAP), RT-PCR can detect the presence of 500-5000 copies of viral RNA/mL near 100 percent of the time. However, pre-analytical factors, such as sample collection technique and the viral load at the collection site, affect the sensitivity and specificity of RT- PCR.17 For example, if most of the viral load is in the lungs, but a nasal swab is collected, then the test may be negative because the viral load is not present at the nasal collection site.
In addition to assays that detect a single virus, some assays detect multiple pathogens. Having an assay that detects more viruses decreases the amount of testing a patient may undergo to receive a diagnosis.18 Rather than performing a test for influenza, respiratory syncytial virus (RSV), and COVID-19 separately with multiple different patient specimens, some assays require one specimen from the patient and can detect a positive result in less than an hour. A disadvantage of performing this testing is that the reagents are in short supply due to the high demand to detect COVID-19 infection.
A variety of assays and tabletop analyzers were approved for emergency use by the U.S. Food and Drug Administration (FDA) during the pandemic.
Tabletop analyzers use isothermal technology to rapidly replicate the viral RNA. An acidic solution is heated to 132.8 Fahrenheit, which damages the viral envelope of COVID-19, exposing the RNA for replication.
Testing advantages include the fast turnaround time and portability, making it ideal for any decentralized lab location.19 A disadvantage to this type of test is seen in the high false-negative rate. In a 2020 research study conducted by the Cleveland Clinic, 239 known COVID-19 specimens were tested on a tabletop analyzer, but only 85.2% of the specimens were considered positive. The desired sensitivity of a rapid test is 95%.20
The same isothermal nucleic acid technology can be used to replicate and differentiate between IAV and IAB nucleic acids.19, 20, 21, 22
Antigen Testing
Antigen testing is another method that can be used to diagnose COVID-19 and influenza infections. Antigen testing is a rapid testing method that detects the presence of viral antigens in the host. This method is not as sensitive or specific as molecular testing. This testing method is a useful tool when screening for COVID-19 or influenza. The high specificity is the ability of the test to determine the absence of an infection. This testing method is cost-effective and only requires a small cartridge, or tabletop analyzer, rather than the large analyzer that was discussed previously. This testing method produces results for COVID-19 influenza A and B in minutes.23, 24
One example of antigen testing for detection of the influenza virus is a rapid influenza diagnostic test (RDIT). This testing method detects viral antigens in 10-15 minutes.25 The tests identify IAV and IBV nucleoprotein antigens present in respiratory specimens.26
Serological testing
Serological testing can also be used to detect a COVID-19 infection. Rather than replicating a target genetic sequence in PCR, serological testing detects the presence of antibodies produced as an immune response by a person infected with a virus. Immunoassays for qualitative and quantitative detection of IgG or IgM antibodies to SARS-CoV-2 in serum are available.27 Serological testing can be used for influenza infections as well; however, the assays are not recommended for clinical diagnosis of IAV or IBV, as the diagnosis cannot be reliably interpreted since testing requires paired acute and convalescent sera collected 2-3 weeks apart for accurate testing.25 This does not allow for timely or rapid detection of IAV or IBV.26 Serological testing is relevant to the public health field and is a valuable screening method.26, 28
Correlated lab findings
While diagnostic testing is necessary to accurately diagnose COVID-19, other laboratory tests help providers monitor patients with COVID-19. Among these tests are Interleukin- 6 (IL-6), D-dimer, Ferritin, Procalcitonin (PCT), and NT- proB- type Natriuretic Peptide (NT-proBNP).15
IL-6 is a predictor of hyper-inflammation and respiratory failure in COVID-19 patients.15 In a meta-analysis conducted in 2020, the “mean IL-6 levels were more than three times higher in patients with complicated COVID-19 compared with those with noncomplicated disease.”29 The increase in IL-6 is related to the “cytokine storm” that occurs in patients with severe COVID-19. The damaging immune response that cytokines promote can lead to acute respiratory distress syndrome.15 Therefore, an increased IL-6 is a predictor for a negative outcome for a patient with COVID-19, such as the need for a respirator.
D-dimer is another laboratory test that can be useful in managing patients with COVID-19. D-dimer measures degradation products from fibrinolysis after clot formation. In COVID-19 patients, D-dimer levels were reported to be elevated in severe cases. A 2020 study conducted by Berger et al. concluded that considering variables such as hypertension, sex, age, and weight, patients with an elevated D-dimer have an increased association with COVID-19 infection.30 Another study found that of the 540 COVID-19 patients that were hospitalized, 69.4% of those COVID-19 patients had elevated D-dimer levels.15
Ferritin is an intracellular blood protein that contains iron and can be measured to determine the amount of iron that is stored in the body. Ferritin levels may aid COVID-19 diagnosis because patients with severe COVID-19 infection show an increase in ferritin levels of hyperferritinemia. Hyperferratinemia among COVID-19 patients is associated with the cytokine storm that takes place during infection. The cytokine storm increases the amount of pro-inflammatory cytokines. The production of these pro-inflammatory cytokines increases inflammation in the body. When inflammation increases, ferritin levels can indicate the degree of inflammation in a patient.
In a study by Gomez-Pastora et al., ferritin levels of patients in Wuhan, China, with non-severe and severe infection were analyzed. The findings for the study showed that patients with a non-severe infection had normal ferritin levels; whereas, patients with a severe infection that survived had ferritin levels >800 ug/L (reference range is 30-400 ug/L). Those that did not survive the infection had ferritin levels that reached 1400ug/L.31
PCT is the precursor of the hormone calcitonin (CT). In bouts of systemic inflammation, PCT is broken down, resulting in elevated PCT in the serum. For elevation of PCT, there must be severe inflammatory stimuli present. An increased PCT is associated with a 5-fold increase in the risk of severe COVID-19 infection.32 A 2020 retrospective cohort study was done with 95 patients with confirmed COVID-19 infections in Union Hospital of Tongji Medical College in Wuhan, China. Of the patients that were studied, 62 had a moderate infection, 21 had a severe infection, and 12 patients were in critical condition. Six of the critical patients died due to COVID-19 infection. In the study, the mean values of PCT were determined for the varying disease severity. In the moderate infection group, the PCT measured 0.05 ± 0.05ng/mL (reference range: <0.15ng/mL). In the severe group, the PCT was 0.23 ± 0.26ng/mL. Lastly, the critical group PCT levels were 0.44 ± 0.55ng/mL.32 The retrospective study was able to correlate the severity of COVID-19 infection with increased PCT levels. The findings of the study detailed that the higher the PCT values in patients with COVID-19 infection, the more likely the patient will have a more severe disease state.
NT-proBNP is a nonactivated pro-hormone that is released in response to changes of pressure inside of the heart and secreted when there is an increase in myocardial wall stress.33 An increase in NT-proBNP is typically associated with heart failure but is also seen in patients with COVID-19. A 2020 retrospective study conducted by Gao Lei and Jiang Dan et al. evaluated the value of NT-proBNP in patient prognosis. In the study, lab data of 102 patients with severe COVID-19 infection at Hubei General Hospital in Wuhan, China, was used in an analysis of NT-proBNP. In the study, a NT-proBNP level of 88.64pg/mL (reference range: 0-74 years of age is 125pg/mL) was the cutoff level to predict in-hospital death in patients with COVID-19 infection. The cutoff value of 88.64pg/mL had 100% sensitivity and 66% specificity. Patients with NT-proBNP levels >88.64pg/mL were also found to have a higher risk of death than patients with a NT-proBNP <88.64pg/mL.34 The retrospective study concluded that the levels of NT-proBNP may be able to determine the risk factor of in-house death for patients with severe COVID-19 infection.
Vaccines
Many COVID-19 vaccination options are under clinical trials or approved for emergency use by the FDA. Table 1 shows some of the vaccination options for COVID-19 around the world. In Table 1, the mechanism of action, dosage, efficacy, storage temperature, and approval status are discussed. The efficacy is the ability of the vaccinations to reduce the risk of contracting the targeted virus. The higher the efficacy of the vaccination, the lower the risk is of contracting the intended virus. Table 2 discusses the most effective COVID-19 and influenza vaccinations currently available.
Viral mutations and vaccinations
It is necessary to understand the structure of SARS-CoV-2 and influenza, due to the challenges created by mutations for vaccination development. Regarding the influenza virus, antigenic drift and antigenic shift are the main challenges associated with vaccination efficacy. The main challenge of SARS-CoV-2 is the emergence of variant strains. The influenza virus undergoes mechanisms called antigenic drift and antigenic shift, which allow the H and N glycoproteins on the surface of influenza to change their expression. When the H and N glycoprotein change due to amino acid substitution or mutation, there is the potential for a new strain of the influenza virus. An example of this is H5N1, or bird flu, which has the H5 glycoprotein expressed, changing the glycoprotein’s properties. In the example above, the H protein in H5N1 would function differently than H1N1 or swine flu.
Antigenic drift and antigenic shift are challenges for vaccination development. As a result, the WHO makes its recommendations for the influenza strain predicted to be the most prevalent six to eight months before the influenza season. The six- to eight-month window is necessary for vaccine development, manufacturing, and distribution.44 However, by the influenza season, antigenic shift or antigenic drift can occur, making the manufactured vaccination ineffective against the virus. In Table 2, the vaccine efficacy for influenza virus ranges from 44-66%. The changes H and N undergo due to antigenic drift and shift explain the range of vaccine efficacy.
The target for vaccine development for SARS-CoV-2 is the Spike glycoprotein on the surface of the virus. In Table 2, the vaccine efficacy for SARS-CoV-2 ranges from 62-95%. The prevalent SARS-CoV-2 variants have Spike glycoprotein mutations. When mutations of the target protein occur, the vaccinations undergo further clinical trials to ensure that the vaccine will be effective against new variants. There are multiple SARS-CoV-2 variants of concern, such as the B.1.351 (Beta), B.1.617 (Delta), and P.1.1 (Gamma) variants.45 A variant of concern can have more severe infection, reduced vaccine effectiveness, or increased transmissibility. The B.1.6.17.2, or Delta variant, is now the most concerning variant of SARS-CoV-2.45
Conclusion
There are many challenges posed by SARS-CoV-2 and the influenza virus regarding diagnosis and prevention. The structure, symptoms, transmission, and rate of mutation of these viruses contribute to these challenges. The spike glycoprotein is the target of vaccination development of SARS-CoV-2. Hemagglutinin and Neuraminidase are the targets for vaccination development of IAV and IBV. Both COVID-19 and influenza have some similar symptoms, such as cough, fever, fatigue, and sore throat; however, the hallmark symptom of COVID-19 is shortness of breath. Both viruses can be transmitted through respiratory droplets, or aerosols, and have varying times of survivability on surfaces, including doorknobs and counters. The lack of proofreading associated with RNA polymerase lends itself to mutations, which can create variants of influenza and COVID-19.
Variants of influenza have a differing expression of H and N glycoproteins. An example of this is H1N1 or swine flu. The only way to confirm a diagnosis of COVID-19 or influenza is confirmatory laboratory testing, which should be considered in conjunction with the presenting symptoms to diagnose COVID-19 and influenza.
Molecular testing is the most sensitive and specific method to confirm COVID-19 and influenza infection. Serological testing is used to detect a previous infection of COVID-19 because serological testing detects the presence of antibodies that are produced by the host as a response to infection. Serological testing is not recommended to detect recent influenza infection because serological testing cannot be reliably interpreted due to the rapid mutation rate of influenza. Antigen testing is a rapid method that can determine the presence of COVID-19 or Influenza infection, but antigen testing is less sensitive and specific than the molecular method of RT-PCR.
Currently, in the United States, three vaccines are offered to the public, Moderna, Pfizer-BioNtech, and Johnson & Johnson / Janssen. Moderna and Pfizer are both two-dose mRNA vaccines; whereas, Johnson & Johnson’s Janssen vaccine is a single dose viral vector vaccination.
References
- Potter CW. A history of influenza. Journal of Applied Microbiology. 2001;91(4): 572-579. https://doi/full/10.1046/j.1365-2672.2001.01492.x.
- Up to 650 000 people die of respiratory diseases linked to seasonal flu each year. World Health Organization. 2017. https://www.who.int/news/item/13-12-2017-up-to-650-000-people-die-of-respiratory-diseases-linked-to-seasonal-flu-each-year. Published December 13, 2017. Accessed April 27, 2021.
- Mittal A, Manjunath K, Ranjan RK, Kaushik S, Kumar S, Verma V. COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020 Aug; 16(8): e1008762. doi: 10.1371/journal.ppat.1008762.
- Ryding S. What is a positive-sense single-stranded rna (+ssrna) virus? https://www.news-medical.net/health/What-is-a-Positive-Sense-Single-Stranded-RNA-(2bssRNA)-Virus.aspx. News Medical.Net.Published March 9, 2021. Accessed March 14, 2021.
- Siu YL, Teoh KT, Lo J, et al. The m, e, and n structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol. 2008 Nov;82(22):11318-30. doi: 10.1128/JVI.01052-08.
- U. Pradesh PDDU, Lee C, Faruku. Bande SSA, et al. Coronavirus envelope protein: Current knowledge. Virology Journal. 2019; https://virologyj.biomedcentral.com/articles/10.1186/s12985-019-1182-0.
- Cornelis AM, de Haan P, Rottier JM. Molecular interactions in the assembly of Coronaviruses. Advances in Virus Research.2005;64: 165-230. https://doi.org/10.1016/S0065-3527(05)64006-7.
- Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008 Sep 12; 26(Suppl 4): D49–D53. doi: 10.1016/j.vaccine.2008.07.039.
- Higgins D, Eckert A. Details - public health image library(phil). Centers for Disease Control and Prevention. https://phil.cdc.gov/Details.aspx?pid=23313. Published 2020. Accessed July 28, 2021.
- Jung HE, Lee HK. Host protective immune responses against influenza a virus infection. Viruses 2020, 12(5), 504; doi.org/10.3390 /v12050504.
- Centers for Disease Control and Prevention. Antigenic characterization. https://www.cdc.gov/flu/about/professionals/antigenic.htm. Published October 15, 2019. Accessed April 12, 2021.
- Centers for Disease Control and Prevention. How covid-19 spreads. https://www.cdc.gov/coronavirus/2019-ncov/transmission/index.html. Published January 7, 2021. Accessed March 14, 2021.
- Centers for Disease Control and Prevention. Emergence of SARS-CoV-2 B.1.1.7 Lineage – United States, December 29, 2020–January 12, 2021. https://www.cdc.gov/mmwr/volumes/70/wr/mm7003e2.htm. Published January 21, 2021. Accessed April 23, 2021.
- Barr J, Fearns R. How rna viruses maintain their genome integrity. J Gen Virol. 2010 Jun;91(Pt 6):1373-87. doi: 10.1099/vir.0.020818-0.
- Jones J. How lab diagnostics can support COVID-19 patient management. Medical Laboratory Observer. https://www.mlo-online.com/continuing-education/article/21202520/how-lab-diagnostics-can-support-covid19-patient-management. Published December 22, 2020. Accessed July 23, 2021.
- Buckingham L, Flaws M. Molecular Diagnostics Fundamentals, Methods, & Clinical Applications. Philadelphia, PA: F.A. Davis; 2007.
- Yohle S. How good are covid-19 (sars-cov-2) diagnostic pcr tests? College of American Pathologists. https://www.cap.org/member-resources/articles/how-good-are-covid-19-sars-cov-2-diagnostic-pcr-tests. Published 2020. Accessed March 14, 2021.
- Xpert Xpress FLU. Cepheid. https://www.cepheid.com/en_US/tests/Critical-Infectious-Diseases/Xpert-Xpress-Flu. Published 2016. Accessed April 4, 2021.
- Abbott AdviseDx SARS-CoV-2 IgM. FDA. https://www.fda.gov/media/142940/download. Published 2020. Accessed 2021.
- Stein R. Study raises questions about false negatives from quick covid-19 test. NPR. https://www.npr.org/sections/health-shots/2020/04/21/838794281/study-raises-questions-about-false-negatives-from-quick-covid-19-test. Published April 21, 2020. Accessed April 4, 2021.
- ID NOW COVID-19. Abbott. https://www.globalpointofcare.abbott/en/product-details/id-now-covid-19.html. Published 2021. Accessed April 4, 2021.
- ID now influenza a & b 2. Abbott. https://www.globalpointofcare.abbott/en/product-details/id-now-influenza-ab-2.html. Published 2019. Accessed April 4, 2021.
- XpertXpress SARS-CoV-2/Flu/RSV. Cepheid. https://www.cepheid.com/en_US/package-inserts/1913. Published 2021. Accessed April 4, 2021.
- QuickVue SARS Antigen Test. Quidel. https://www.quidel.com/immunoassays/quickvue-sars-antigen-test. Published 2021. Accessed April 22, 2021.
- Centers for Disease Control and Prevention. Overview of influenza testing methods.https://www.cdc.gov/flu/professionals/diagnosis/overview-testing-methods.htm. Published August 31, 2020. Accessed August 16, 2021.
- Centers for Disease Control and Prevention. Rapid influenza diagnostic tests. https://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm. Published October 25, 2016. Accessed August 16, 2021.
- Centers for Disease Control and Prevention. Serology testing for Covid-19 at CDC. https://www.cdc.gov/coronavirus/2019-ncov/lab/serology-testing.html. Accessed August 15, 2021.
- Sofia Influenza A+B FIA. Quidel. https://www.quidel.com/immunoassays/rapid-influenza-tests/sofia-influenza-fia. Published 2021. Accessed April 22, 2021.
- Grifoni E, Valoriani A, et al. Interleukin-6 as prognosticator in patients with COVID-19. J Infect. 2020 Sep;81(3):452-482. doi: 10.1016/j.jinf.2020.06.008.
- Berger J, et al. Prevalence and outcomes of d-dimer elevation in hospitalized patients with covid-19. Arterioscler Thromb Vasc Biol. 2020 Oct;40(10):2539-2547. doi: 10.1161/ATVBAHA.120.314872.
- Gómez-Pastora J, Weigand M, Kim J, et al. Hyperferritinemia in critically Ill COVID-19 patients - Is FERRITIN the product of inflammation or a Pathogenic mediator? Clin Chim Acta. 2020 Oct;509:249-251. doi: 10.1016/j.cca.2020.06.033.
- Hu R, Han C, Pei S, Yin M, Chen X. Procalcitonin levels IN COVID-19 Patients. Int J Antimicrob Agents. 2020 Aug; 56(2): 106051 doi: 10.1016/j.ijantimicag.2020.106051.
- NT-proB-type Natriuretic Peptide (BNP). Cleveland Clinic.https://my.clevelandclinic.org/health/diagnostics/16814-nt-prob-type-natriuretic-peptide-bnp. Published 2019. Accessed March 28, 2021.
- Gao L, Jiang D, Wen X-song, et al. Prognostic value Of NT-proBNP in patients with Severe covid-19. Respir Res. 2020;21(1):83. doi: 10.1186/s12931-020-01352-w.
- Vaccination Provider Fact Sheet | EUA | Moderna COVID-19 Vaccine. ModernaTx. Moderna. https://www.modernatx.com/covid19vaccine-eua/eua-fact-sheet-providers.pdf. Published 2020. Accessed April 22, 2021.
- Food and Drug Administration. Fact Sheet for Healthcare Providers Administering Vaccine (VACCINATION PROVIDERS). https://www.fda.gov/media/144413/download. Published August 12, 2021. Accessed August 15, 2021.
- Johnson & Johnson COVID-19 vaccine authorization by U.S. FDA for Emergency Use. Johnson & Johnson. https://www.jnj.com/johnson-johnson-covid-19-vaccine-authorized-by-u-s-fda-for-emergency-usefirst-single-shot-vaccine-in-fight-against-global-pandemic. Published 2021. Accessed April 22, 2021.
- AstraZeneca. COVID-19 Vaccine AstraZeneca. EMA. https://www.ema.europa.eu/en/documents/product-information/covid-19-vaccine-astrazeneca-product-information-approved-chmp-29-january-2021-pending-endorsementNovavax. PREVENT-19: The Novavax Phase 3 Clinical Trial in the U.S. and Mexico. Novavax. https://www.novavax.com/sites/default/files/2021-01/Novavax-PREVENT-19-Factsheet.pdf. Published 2021. Accessed April 22, 2021.
- Novavax COVID-19 Vaccine demonstrates 90% overall efficacy and 100% protection against moderate and severe disease in prevent-19 Phase 3 trial. Novavax Investor Relations. https://ir.novavax.com/2021-06-14-Novavax-COVID-19-Vaccine-Demonstrates-90-Overall-Efficacy-and-100-Protection-Against-Moderate-and-Severe-Disease-in-PREVENT-19-Phase-3-Trial. Published June 14, 2021. Accessed August 16, 2021.
- Novavax COVID-19 Vaccine demonstrates 90% overall efficacy and 100% protection against moderate and severe disease in prevent-19 Phase 3 trial. Novavax Investor Relations. https://ir.novavax.com/2021-06-14-Novavax-COVID-19-Vaccine-Demonstrates-90-Overall-Efficacy-and-100-Protection-Against-Moderate-and-Severe-Disease-in-PREVENT-19-Phase-3-Trial. Published June 14, 2021. Accessed August 15, 2021.
- U.S. Food and Drug Administration. Seqirus. https://www.fda.gov/media/117022/download. Published 2020. Accessed 2021.
- U.S. Food and Drug Administration. GlaxoSmithKline Biologicals. Package Insert - FLUARIX QUADRIVALENT. https://www.fda.gov/media/79278/download. Published 2020. Accessed April 22, 2021.
- U.S. Food and Drug Administration. MedImmune. Package Insert - FluMist Quadrivalent. https://www.fda.gov/media/83072/download. Published 2019. Accessed April 22, 2021.
- Webster RG, Govorkova EA. Continuing challenges in influenza. Annals of the New York Academy of Sciences. 2014;1323(1):115-139. doi:10.1111/nyas.12462.
- Centers for Disease Control and Prevention. Sars-cov-2 variant classifications and definitions. CDC.gov. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#Concern. Published 2021. Accessed July 28, 2021.
About the Author

Sophia Richards, MLS(ASCP)
A graduate of the Medical Laboratory Sciences Program at Northern Illinois University, Sophia Richards, MLS (ASCP), is as a medical laboratory scientist at OSF St. Anthony Hospital in Rockford, IL.

Masih Shokrani, PhD, MT(ASCP)
is a professor in the Medical Laboratory Sciences Program of Northern Illinois University. He teaches medical immunology, medical diagnostic biochemistry and research.