The significance of serology antibody testing for SARS-CoV-2
The World Health Organization (WHO) declared the coronavirus SARS-CoV-2 disease (COVID-19) outbreak a global pandemic on March 11, 2020.1 This is the third coronavirus to cross species, following the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012, and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-1) in 2003.2 However, SARS-CoV-2 has had a more rapid spread with over 7.2 million confirmed cases and over 400,000 deaths worldwide.3 The United States has nearly 2 million confirmed COVID-19, cases and above 100,000 related deaths.3
As of June 9, 2020, the U.S. Food and Drug Administration (FDA) had provided Emergency Use Authorizations (EUAs) for molecular-based detection and/or diagnosis of COVID-19 to 74 manufacturers and 36 high-complexity laboratories who had developed molecular-based laboratory developed tests.4 Also as of June 9, 2020, the FDA has issued only 19 EUAs for antibody-based detection tests. The FDA has not objected to the development and distribution of serology tests in identifying antibodies for SARS-CoV-2 as long as manufacturers apply for EUA within 10 days of notifying the agency of their intent to market the tests.5
Why is serology testing important for SARS-CoV-2?
Currently, SARS-CoV-2 infection is diagnosed by the detection of the virus via molecular-based techniques, such as real-time reverse transcriptase polymerase chain reaction (RT-PCR) or deep sequencing. However, these tests have a high rate of false negativity. 6-8 These detection systems rely on adequate viral load at the sample collection site.6 SARS-CoV-2 can cause both upper, but predominantly lower, respiratory tract infections. The positivity rate for the different samples ranges from 74.4 percent to 88.9 percent for sputum, 53.6 percent to 73.3 percent for nasal swabs, and a much lower rate in throat swabs collected ≥8 days post-disease onset.9
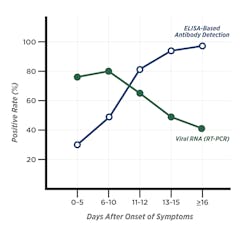
The highest positivity rate has been shown in bronchoalveolar lavage fluid specimens (93 percent).10 However, lower respiratory tract sampling requires both a skilled technician and a suction device. Additionally, false-negative results are observed with RT-PCR methods when the viral replication time-window is missed.6 Liu et al. 2020 showed 11 days’ post symptom onset (PSO) is a transitional time point, when infected patients produce anti-viral antibodies, allowing confirmation of viral infection. The positivity rate for detection of IgM and/or IgG antibodies was >80 percent at this time point and was close to 100 percent after 14 days PSO. In contrast, the positivity rate of RT-PCR was about 60 percent 11 days PSO and began to rapidly decline afterwards (Figure 1).
The overall positivity rate was significantly higher by antibody testing (81.5 percent) compared with RT-PCR (64.3 percent) at day 11 PSO.7 In another study, only 51.9 percent of patients were positive by a single RT-PCR test, but the positivity rate increased to 98.6 percent following and antibody assay on PCR- negative individuals.6 At this stage of the pandemic, a molecular false-negative result could pose a serious threat by allowing infectious patients back into the community, and hindering the efforts to contain virus spread.6 Therefore, a combination of RT-PCR and antibody testing methods would considerably improve diagnostic efficacy, even during early stages.7
What is the significance of serology testing for SARS-CoV-2?
Serological assays aid in understanding the immune responses to SARS-CoV-2 in a dynamic qualitative or quantitative manner and to identify individuals who were infected compared to those who were not. Data from serological tests are essential to determine the immune status of healthcare workers. Those who have developed the antibodies then can be deployed back to frontline work with less risk of illness while limiting virus spread.11 Serological assays are also needed to conduct epidemiologic studies to assess the extent of virus spread in communities and to determine the infection fatality rate. Furthermore, serological assays can identify individuals with the required antibody response against SARS-CoV-2, who can be donors of hyperimmune plasma to treat sick individuals.11,12 Lastly, serological assays can be used to select patients for clinical trials for vaccine or therapy development.2
What are the target antigens for the serology testing for SARS-CoV-2?
The N protein plays an important role in the transcription and replication of viral RNA, packaging the encapsidated genome into virions16 and inhibits the cell cycle process of the host cells.17 The N protein is abundantly expressed during infections and also has high immunogenic activity.8,9,18 Therefore, both N and S protein could be potential targets for the antibody-based detection of SARS-CoV-2.9 However, the N protein homology between SARS-CoV-2 and SARSCoV-1 is 90 percent, compared with the S protein (77 percent), especially the S1 subunit including the RBD (66 percent).
Additionally, the percentage of amino acid identity among other endemic coronaviruses (OC43, HKU1, 229E, NL63) and MERS-CoV was higher for the N protein, compared with the S1 subunit,2 suggesting that serodiagnostics using N protein could potentially demonstrate greater cross-reactivities among the endemic coronaviruses. Jiang et al. 2020 showed the N protein based antibody assays could exhibit a higher false-negative rate compared with the S1 subunit, and that S1 subunit purified from mammalian cells demonstrated the highest performance to distinguish COVID-19 patients from controls.19
On the contrary, the N protein antigen is generally produced in bacteria instead of mammalian cell lines. Therefore, critical human glycosylation would be missing and, in turn, decrease high-affinity binding to antibodies.20 Therefore, the S1 subunit could be the specific target antigen for detecting SARS-CoV-2 antibodies.2
What antibodies are detected against SARS-CoV-2?
In infectious disease, IgM and IgG antibodies are commonly detected using serological tests due to their important role in tackling the viral infection. However, in many respiratory infections, in addition to IgM antibodies, IgA antibodies are produced in high titers during the early phase of the infection.21,22 In general, IgM antibodies have a lower affinity compared to IgA or IgG antibodies, and pose an increased risk of cross-reactivity against antigenically similar epitopes, which are common in coronaviruses.23
During the SARS-CoV-1 infection of 2002, there was simultaneous seroconversion of all immunoglobulin classes (IgM, IgA and IgG),24,25 and similar observations are being reported for the SARS-CoV-2 infection. 6,20,26 Guo et al. 2020 showed that the production of IgA, IgM, and IgG antibodies was positive as early as one-day PSO. IgM/IgA antibodies and IgG antibodies were detected at a median of five and 14 days’ PSO, respectively.
Additionally, during the acute phase (days 1-7, PSO), a higher positivity rate was observed for IgA antibodies (92.7 percent) compared with IgM antibodies (85.4 percent),6 indicating IgA antibodies could serve as a marker for acute infection. In another study, IgA antibodies were detected earlier than IgM antibodies, and the IgA values peaked at 20-22 days with persistently higher levels compared with IgM antibodies throughout the observation period (45 days PSO).
In contrast, IgM levels peaked at 10-12 days and declined after 18 days.27 Jin et al. 2019 showed as the interval from RT-PCR confirmation to serological detection increased, the IgM positivity rate (75 percent) increased initially but subsequently declined (<50 percent). IgG positivity increased to 100 percent and remained stable thereafter. IgG levels were consistently higher than IgM levels,8 suggesting IgG antibodies persist longer in the body, and could contribute to long-term immune memory against SARS-CoV-2.28.Does the presence of antibodies indicate if someone is immune against SARS-CoV-2 reinfection?
The presence of antibodies alone does not indicate that the person is immune against potential reinfection. It is important to determine whether the antibodies can confer protective immunity long-term, providing virus-neutralizing antibodies that block viral infection and help in clearing viral infection.14,28 Therefore, neutralizing antibodies play an important role in resolving viral infection. Plaque reduction neutralization tests (PRNT) in vitro are considered the gold standard for determining the ability of antibodies to neutralize virus and prevent viral replication.28 Neutralizing antibodies primarily target S protein in coronaviruses, in particular the S1 subunit and the RBD contained within the S1 domain, preventing viral entry into the host cell.29
Studies from 2002 SARS-CoV-1 and 2012 MERS-CoV have reported neutralizing antibodies target the S protein, predominantly S1 or the RBD, and to a lesser extent S2,30 and similar observations are emerging in SARS-CoV-2. Neutralizing antibody responses against SARS-CoV-1 begin to develop by week two, and most patients develop neutralizing antibodies by week three.14 Okba et al. 2020 compared the performance of different ELISAs with PRNT assays to determine their ability to detect neutralizing antibodies to SARS-CoV-2. They found good correlation between antibody results in different ELISAs and the neutralizing test.
The strongest correlation was observed with the EUROIMMUN IgA ELISA for infection reduction rates of >90 percent (PRNT90), suggesting the ability of EUROIMMUN ELISAs to detect SARS-CoV-2-specific antibodies.2 However, it is currently unknown whether the presence of antibodies to SARS-CoV-2 is sufficient to confer protective-immunity in vivo in infected individuals. SARS-CoV-2 is a novel pathogen, and there is no information regarding the duration or long-term effectiveness of these antibodies in recovered patients. SARS-CoV-1 specific antibodies became undetectable in 91 percent of patients after six years of infection,31 indicating longitudinal studies are required to further understand the potential role of SARS-CoV-2 specific antibodies in patients.
Conclusion
Detection of antibodies against SARS-CoV-2 virus complements viral testing. Antibody detection in combination with RT-PCR expands the detection window of SARS-CoV-2 infection and minimizes false-negative RT-PCR testing. It would provide valuable information to public health authorities making decisions to relax physical distancing measures and reopen businesses across the country. Antibody testing will help determine the extent of SARS-CoV-2 spread in communities. However, there are still questions that need to be answered such as how long would the antibodies last and if they are truly protective?28
It is important to understand the antibody kinetics in SARS-CoV-2 patients. Studies have reported both IgA and IgM antibodies can be detected earlier than IgG antibodies.20,32 Additionally, IgA and IgM levels peak early post infection, but IgA is more robust and longer lasting. IgG levels peak at 21-25 days PSO.20 However, further studies are required to understand the long-term kinetics of these antibodies in infected individuals. Additionally, whether these antibodies can provide protection from reinfection and what antibody titers are required for protective immunity are yet to be determined. Furthermore, scientists are working on developing an effective vaccine, but the timeframe for its availability is unclear.
With many states in the U.S. looking to re-open businesses, it is important to be vigilant and continue practicing social distancing and good hygiene practices until the role of antibodies against the SARS-CoV-2 virus is better understood.
REFERENCES
- WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. Published 2020. Updated March 11, 2020. Accessed April 24, 2020.
- Okba NMA MM, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7).
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html. Published 2020. Updated June 10, 2020. Accessed June 10, 2020.
- Coronavirus Disease 2019 (COVID-19) Emergency Use Authorizations for Medical Devices. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#covid19ivd. Published 2020. Updated June 9, 2020. Accessed June 10, 2020.
- Policy for Coronavirus Disease–2019 Tests During the Public Health Emergency (Revised). https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-public-health-emergency-revised. Published 2020. Updated 05/11/2020. Accessed 06/10, 2020.
- Guo L, Ren L, Yang S, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin Infect Dis. 2020. DOI: 10.1093/cid/ciaa310
- Liu L, Liu W, Wang S, Zheng S. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. BMJ (In Press). 2020.
- Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020; 94: 49-52.
- Liu W, Liu L, Kou G, et al. Evaluation of Nucleocapsid and Spike Protein-based ELISAs for detecting antibodies against SARS-CoV-2. Journal of Clinical Microbiology. 2020: 58(6):e00461-20.
- Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323(18):1843-1844.
- Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nature Medicine. 2020.
- To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565-574.
- Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273.
- Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nature Reviews Immunology. 2020.
- Woo PCY, Lau SKP, Wong BHL, et al. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. Journal of Clinical Microbiology. 2005;43(7):3054-3058.
- Chen C-Y, Chang C-K, Chang Y-W, et al. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 2007;368(4):1075-1086.
- Cui L, Wang H, Ji Y, et al. The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells. J Virol. 2015;89(17):9029-9043.
- Che X-Y, Qiu L-W, Pan Y-X, et al. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. Journal of Clinical Microbiology. 2004;42(6):2629-2635.
- Jiang H-w, Li Y, Zhang H-n, et al. Global profiling of SARS-CoV-2 specific IgG/ IgM responses of convalescents using a proteome microarray. medRxiv (preprint). 2020:2020.2003.2020.20039495.
- Ma H, Zeng W, He H, et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by chemiluminescence immunoanalysis. medRxiv (preprint). 2020:2020.2004.2017.20064907.
- Renegar KB, Small PA, Jr., Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol. 2004;173(3):1978-1986.
- Robert-Gangneux F, Darde ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. 2012;25(2):264-296.
- Meyer B, Drosten C, Muller MA. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175-183.
- Hsueh PR, Huang LM, Chen PJ, Kao CL, Yang PC. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10(12):1062-1066.
- Woo PC, Lau SK, Wong BH, et al. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin Diagn Lab Immunol. 2004;11(4):665-668.
- Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020:2020.2003.2017.20037713.
- Padoan A, Sciacovelli L, Basso D, et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clinica Chimica Acta; International Journal of Clinical Chemistry. 2020; 507: 164-166.
- Gigi Gronvall PNC, PhD; Amanda Kobokovich, MPH; Rachel West, PhD; Kelsey Lane Warmbrod, MS, MPH; Matthew P. Shearer, MPH; Lucia Mullen, MPH; Tom Inglesby, MD. Developing a National Strategy for Serology (Antibody Testing) in the United States. Johns Hopkins Center for Health Security; April 22, 2020 2020.
- Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nature Communications. 2020;11(1):2251.
- Jiang S, Hillyer C, Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends in Immunology. 2020:5.
- Tang F, Quan Y, Xin Z-T, et al. Lack of Peripheral Memory B Cell Responses in Recovered Patients with Severe Acute Respiratory Syndrome: A Six-Year Follow-Up Study. The Journal of Immunology. 2011;186(12):7264.
- Lee CY-P, Lin RTP, Renia L, Ng LFP. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Frontiers in Immunology. 2020;11.
About the Author

Iswariya Venkataraman, PhD
serves as Scientific Affairs Lead, EUROIMMUN U.S. She’s currently a member of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Private Partner Scientific Board (PPSB), an independent, pre-competitive forum for study-related scientific exchange in the field of neurodegeneration. During her doctoral studies in neuroscience, she identified novel auto-antigens in autoimmune hyper-excitability disorders.

Maite Sabalza, Ph.D.
is the Scientific Affairs Manager at EUROIMMUN US. Her academic background is in infectious diseases and diagnostics. In her role at EUROIMMUN US, she establishes relationships with key opinion leaders (KOLs) and supports the team with commercial activities including scientific collaborations, scientific marketing, and business development of diagnostics.

Stanley J. Naides, MD, FAP, FACR
Director of Scientific Affairs, EUROIMMUN US. Dr. Naides is a clinician-scientist, boarded in internal medicine and rheumatology with a track record of research in viral pathogenesis and laboratory medicine, and of leadership in esoteric laboratory services in immunology.