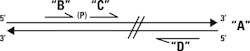The basic concept of the polymerase chain reaction (PCR) has frequently been modified since its inception some three-and-a-half decades ago. One such modification which the reader may encounter is where the assay incorporates some form of obligate DNA ligase step as an addition to the base reaction. Reactions of this type are referred to as ligation-mediated PCR; however, this is a rather broad umbrella term under which a number of quite different functional guises exist. In this month’s Primer, we’ll look at what a couple of these methods are and where the reader might encounter them.
The DNA ligase: the tie that binds
First let’s review what DNA ligase is and what reaction it catalyzes. Recall that DNA is double-stranded, with each strand having an intrinsic polarity defined by the 3′ and 5′ ends of the sugar segments which alternate with phosphate groups to make up the backbone of each strand. The paired strands are “anti-parallel,” meaning their polarities face opposite directions; if viewed from the side, one strand of the helix would have its 3′ end to the right, and the other strand would have its 3′ end to the left.
A nick in a strand is any place where there’s a missing covalent bond between a sugar and the next adjacent phosphate. Nicks can arise from DNA damage, during normal DNA replication, or from the action of nuclease enzymes. Since a nick breaks the continuity of the backbone of one strand, it must be repaired by re-forming the missing covalent bond in order to make the DNA molecule intact again. This job of sealing nicks (specifically, nicks that occur between a sugar 3′ -OH group and the adjacent phosphate attached to the preceding sugar’s 5′ -OH group) is the function of DNA ligase. Using a molecular energy source (which differs depending on the enzyme source organism), DNA ligase reforms the missing covalent bond and the strand is whole again. Two aspects of this are critical:
- The nick to be repaired occurs on a single strand but in the context of a double-stranded molecule.
- The bases of the nicked strand, and particularly those directly flanking the nick site, must be properly base-paired to the opposite (un-nicked) strand.
It’s not hard to imagine why this is: the base pairing is required to hold the two parts (sugar 3′ -OH and the next phosphate) in place for the ligase enzyme active site to catalyze joining them. If either one of these isn’t base-paired down and is flopping about with thermal motion, the reaction geometry doesn’t occur, and no new bond can be made.
Now imagine that the nick in question isn’t one which arose naturally, but instead exists between two synthetic single-stranded DNA pieces which are complementary to a single-stranded target region. If we have an assay where those two fragments must be joined into a single longer fragment to create a signal, then we can use a preliminary ligase-mediated step to test for the existence of their complement. That is, if we want to know whether strand “A” with a defined sequence exists in our sample, we can mix in the two short, adjacent, theoretically complement strands “B” and “C” with their “nick” being next to the base of interest. They will hybridize next to each other and form a substrate for DNA ligase activity only when a perfect match occurs, and are particularly sensitive to disruption from any mismatch near the nick—like a single nucleotide polymorphism (SNP). If and only if a perfect match occurs, B and C get ligated into a single longer
molecule “B+C.”
In the nick of time: detecting ligation
How do we detect that B+C was formed? One way is to imagine that, if together, B+C form one primer of a traditional PCR primer set (call the other “D,” with it being of similar length and annealing temperature to B+C). If we now run a PCR with a primer annealing temperature designed for the full-length B+C/D product, we’ll only get a product if B+C was formed and thereby created the B+C composite primer. (Primers B and C on their own will have much lower annealing temperatures than B+C, so if they have not ligated, they’re relegated to being inactive bystanders in the PCR). By adding on this pre-PCR ligation test step, we’re able to test very specifically for any nucleotide changes in the template at the B+C ligation region.
Why not just skip the ligation fuss, and use a premade B+C primer sequence? In theory it should show less hybridization affinity to a sequence with any mismatch than to a perfect complement target. In reality, however, if it’s a single nucleotide in the middle of a primer, mismatches don’t always destabilize priming enough to disrupt a small number of amplicons being formed. Of course, once formed, by nature of the PCR process, these serve as perfect templates for the subsequent round, and your hopefully SNP-specific longer B+C primer probably doesn’t end up in reality as SNP-specific as you’d hoped.
Why doesn’t the ligase in the reaction tube go about randomly ligating products every cycle and just make a huge mess? There’s a good answer for that, too. We use a non-thermostable ligase enzyme, such that it’s permanently denatured and its function is destroyed by the first high-temperature opening denaturation of the classical PCR phase of the test. Conveniently, most ligases are not thermostable, and most ligation reactions have optimal reaction temperatures well below any of the thermal steps in a PCR, so no fancy enzyme engineering is required for this.
There are other ways to detect that B+C was formed. We can just run the ligation portion of the above, then denature the material and use a method such as capillary electrophoresis or mass spectrometry to look for the B+C product, particularly if either B or C, or both, carry appropriate mass or fluorophore tags. Regardless of the method used to detect that B+C was formed, these types of assays are probably the most common example of ligation-mediated PCR the average laboratorian will encounter and are known more specifically as oligonucleotide ligation assays.
Amplified fragment length polymorphism
A completely different type of ligation-mediated PCR is one which readers may have encountered if they had a need to do microorganism genetic typing or “fingerprinting,” such as for tracking epidemiology. An approach called amplified fragment length polymorphism (AFLP) can be used here. In this assay type, purified test organism DNA is collected from a culture and it’s subjected to digestion with a restriction enzyme (or sometimes more than one, but for sake of argument we’ll only consider one). Restriction enzymes are ones which recognize specific, usually 4, 6, or 8 base palindromic sequences in DNA, and create a pair of nicks at the site—one in each strand. This creates a full double-stranded break, usually offset by a few bases between strands to create a staggered or “sticky” end. This restriction digest is run for a significant excess of time, to ensure that practically all cut sites are indeed cut.
The result is that the originally intact organism DNA sample is now cut into a huge number of smaller pieces. Different strains of an organism will have at least some sequence differences such as insertions or deletions (which will change the size of some of the end product fragments) or may have nucleotide substitutions which create or destroy restriction enzyme recognition sites relative to the reference strain. Thus, the pool of fragments created from the complete restriction digestion of the genome of two strain variants will have a finite number of differences.
How can these be detected? If we just run the full digest out on an agarose gel, it will look like a big smear in either case and the few differences will be hidden under the literally millions of fragments of different sizes present (and in most cases, identical between the strains). To get around this, we take our restriction digest product—with known sticky ends—and we add in large quantities of a short double-stranded oligonucleotide adapter molecule with a matching sticky end. When we now add DNA ligase, these adapters can anneal to and then ligate to put known, defined sequence “caps” on all of the fragment types present.
Now what we can do is use this as a template to PCR from, using PCR primers which match the end caps, since we now know these flank all of the target genome fragments. Just doing that would be kind of pointless, though, as it would just replicate all of what’s already a far too jumbled mess. We want to find a way to select out and amplify just a very few representative fragments. To do this, we add a few nucleotides on the 3′ end (inside) of the cap-matching section of the primer. This PCR primer will then only productively anneal to those genomic fragments which have ligated on caps, and whose few terminal nucleotides just happen to be the complement of the primer 3′ addition. Each addition of one 3′ direction nucleotide to the defined cap sequence primer causes a roughly 16-fold decrease in the number of molecules it can match (there are four choices per nucleotide, so a ¼ chance it fits; since it happens at both ends, 1/16). A three-nucleotide addition means only roughly one out of 4,096 fragments is now amplified by PCR—a small enough fraction that the PCR now generates a visually distinctive banding pattern or “fingerprint” when analyzed by gel electrophoresis. Changes to the size of any of these selected fragments constitutes a differentiable genetic marker allowing for samples (strains) to be distinguished—all without having to know anything about the actual DNA sequence of the organisms to be separated, and requiring only the most basic molecular lab equipment.
The two above examples certainly aren’t it for the forms in which ligation-mediated PCR can be found; they are, however, probably the two most common forms the reader is likely to encounter, and provide a useful framework from which to understand most of the other variations on linking PCR to the outcome of a ligation reaction.
John Brunstein, PhD, is a member of the MLO Editorial Advisory Board. He serves as President and Chief Science Officer for British Columbia-based PathoID, Inc., which provides consulting for development and validation of molecular assays.
About the Author

John Brunstein, PhD
is a member of the MLO Editorial Advisory Board. He serves as President and Chief Science Officer for British Columbia-based PathoID, Inc., which provides consulting for development and validation of molecular assays.