Direct-from-whole-blood testing will speed sepsis diagnosis
Amid the many complexities involved in the management of patients with sepsis, the key question is simple: how soon can the correct treatment be initiated? Sepsis is a race against time, because survival rates deteriorate by eight percent for every hour of inappropriate treatment.1 If targeted therapy is provided within a few hours of patient presentation, the progression of a bloodstream infection to sepsis can be prevented, reducing the likelihood of health consequences that are always significant and often fatal.
Lee Health’s experience
Laboratorians at Lee Health System in southwest Florida are leading the way by slashing the time to the detection of sepsis-causing pathogens with blood culture-independent, direct-from-whole-blood testing. The new approach and technology enable clinical health professionals to begin appropriate targeted therapy much sooner than they could with traditional blood culture-dependent assays. In a recent study published in the Journal of Antimicrobial Chemotherapy, investigators at Lee Health found that patients with potentially deadly infections received targeted therapy 28 hours faster with direct-from-whole-blood tests.2
Using the FDA-cleared direct-from-whole-blood fungal panel, the lab also contributed to the hospital’s cost stewardship initiatives and drove savings for the hospital, because faster negative results reduced unnecessary antimicrobial therapy by an average of four days and an estimated cost saving of $280 per patient. Further, Lee Health’s clinical investigators presented research on a new bacteria assay at the May 2018 “Making a Difference in Infectious Disease” (MAD-ID) annual meeting. Lee Health reported that “final results were realized within four hours from direct-from-whole-blood testing” and that a positive result was received 20 hours sooner than a positive blood culture result and a negative result was received 122 hours sooner than a negative blood culture result, a statistical significance of p<0.001.3 The authors identified 37 opportunities for antibiotic de-escalation due to the direct-from-whole-blood testing, further bolstering their stewardship program.
Blood culture: uses and limits
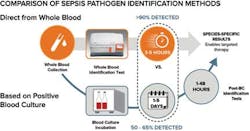
Targeted therapy requires knowing which specific pathogen is at work, and the experience at Lee Health represents a departure from the conventional way of identifying sepsis-causing pathogens in the bloodstream. There have been many important advances over the years to improve the blood culturing process and speed up the post-blood culture tests for identification and antimicrobial susceptibility testing. However, these tests remain limited by the necessarily long time to positivity for a blood culture result, which can be more than 24 hours because blood culture requires growing pathogen cells. In addition, blood culture often misses 35 percent to 50 percent of the organisms for the first blood draw.4,5 The number of cells present after a blood culture positive are typically in the 10,000 to 1,000,000 CFU/mL range, which is a more than 1,000-fold increase in clinically relevant levels of sepsis-causing pathogens in the blood (1-15 CFU/ml). The wait for blood culture positivity, if the cells adequately grow at all, and the time needed for subsequent species identification force clinicians to delay targeted therapy during a critical window of time when a patient is suspected of sepsis.
The unavoidable delays in getting blood culture results have also driven an increase in the use of empiric, broad-spectrum therapy that is based on protocols and probabilities. The push for rapid empiric therapy has saved countless lives in the absence of detailed infection information, but it is still flawed for many clinicians, laboratorians, pharmacists, and patients. Studies have demonstrated that up to 40 percent of patients with bloodstream infections received ineffective empiric therapy.6The consequences of overuse of unnecessary antimicrobials are significant, from the rise of frightening drug-resistant superbugs identified by the Centers for Disease Control and Prevention (CDC) to immediate patient health impacts such as toxicity. The need to get answers before blood culture has been demonstrated by the increasing use of biomarkers such as β-d-glucan and procalcitonin (PCT) that attempt to assist in more rapid therapeutic decisions, especially in a critical care or emergency setting. These tests may provide a helpful indication of infection in certain settings, but there are still significant gaps and clinical limitations due to unreliable accuracy and no species-specific information.
Enter whole-blood diagnostics
A new breakthrough in management for patients suspected of sepsis and bloodstream infections must be driven by the lab through rapid identification of infection-causing species without the wait for blood cultures. The lab is a pivotal member of any critical care team, and it can take patient care to a new level: rapid results from the lab can improve the patient treatment pathway by augmenting existing algorithms to enable treating physicians to make a targeted attack on infections. There are numerous targeted potential pharmaceutical options, but they require specific information from the lab on the infection to provide full value.
In 2014, the FDA cleared the first direct-from-whole-blood test, which provides fungal species identification results in three to five hours.7 In clinical use, rapid identification has enabled targeted therapy on Day 0 and delivered cost savings to the pharmacy budget, as reported in peer-reviewed studies published over the last year.
A bacteria panel, cleared by the FDA in May 2018, identifies five of the most common and deadly sepsis-causing species of bacteria: Enterococcus faecium, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus.8 In a prospective pivotal trial that included 11 centers and more than 1,400 subjects, test results were achieved 2.5 days faster than with blood culture with excellent accuracy, including overall average sensitivity of 90 percent and overall average specificity of 98 percent. In addition, 69 patient infections were detected by the new test that were missed by a blood culture that was run at the same time. The technology enabling this advance combines magnetic resonance with advances in nanotechnology to measure how water molecules react in the presence of magnetic fields. In addition to the manufacturer of the cleared bacterial and fungal tests, a number of diagnostics companies, both large and small, have recognized the clinical value of the approach and are working to enter the emerging direct-from-whole-blood market.
Looking ahead
It will likely be only a matter of time before whole-blood diagnostics become part of the standard of care for patients suspected of sepsis and bloodstream infections. The opportunities for both improved patient care and cost savings are just too great to be ignored. A major milestone for this advance will be the addition of such diagnostics into sepsis protocols—running these tests at the same time as cultures and lactates are run. Indeed, this is already happening in some hospitals.
Cost pressures for labs are ever-increasing, so the economic justification for any new test must be rigorous. In the field of sepsis-related infections, however, the potential to reduce hospital and intensive care unit (ICU) length of stay, while reducing the use of drugs, is compelling. Importantly, there are also potential CPT reimbursement codes for direct-from-whole-blood tests in the outpatient setting. The value of the new tests will be measured by the impact they will have on the amount of money that hospitals spend for sepsis, which is now the most expensive inpatient cost in the United States.
The complete measure of the new assays, however, will be their impact on the sepsis public health crisis as a whole. The implementation of rapid results and treatment is uniquely positioned to strike a huge blow to sepsis and the hundreds of thousands of lives it takes each year. A shift to more accurate and faster testing can potentially result in thousands of patients going home to their families instead of the tragic, and avoidable, alternative.
REFERENCES
- Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589-1596.
- Patch ME, Weisz E, Cubillos A, Estrada SJ, Pfaller MA. Impact of rapid, culture-independent diagnosis of candidaemia and invasive candidiasis in a community health system. J Antimicrob Chemother. 2018: 73(Suppl_ 4):iv27–iv30.
- Weisz E, Newton EC, Estrada SJ, Saunders MB. Early experience with the T2bacteria research use only (RUO) panel at a community hospital. Poster presentation. MAD-ID 2018.
- Clancy CJ, Nguyen MH. Finding the “missing 50%” of invasive candidiasis: how nonculture diagnostics will improve understanding of disease spectrum and transform patient care. Clin Infect Dis. 2013;56(9):1284-1292.
- Cockerill III FR, Wilson JW, Vetter EA, et al. Optimal testing parameters for blood cultures. Clin Infect Dis. 2004;38(12):1724-1730.
- Buehler SS, Madison B, Snyder SR, et al. (2016). Effectiveness of practices to increase timeliness of providing targeted therapy for inpatients with bloodstream infections: a laboratory medicine best practices systematic review and meta-analysis. Clin Microbiol Rev. 2016:29(1):59-103.
- FDA News Release. FDA allows marketing of the first test to identify five yeast pathogens directly from a blood sample. September 22, 2014.
- T2 Biosystems Press Release. T2 Biosystems receives FDA clearance to market T2Bacteria panel for detection of sepsis-causing pathogens. May 29, 2016.