Rapid biomarker testing for improved clinical decision-making in non-small cell lung cancer
CONTINUING EDUCATION
To earn CEUs, visit www.mlo-online.com under the CE Tests tab.LEARNING OBJECTIVES
1. Discuss the statistics on lung cancer in terms of diagnosis time, prognosis, and therapies.
2. Identify the importance of efficient and timely diagnosis and treatment of lung cancer.
3. List the new types of biomarker tests that are accelerating in the field of lung cancer diagnostics.
4. Discuss the capabilities that the biomarker tests provide and the information they can give to physicians.
One devastating aspect of lung cancer is that it can develop slowly and remain undetected for years before becoming symptomatic. By the time lung cancers are discovered and diagnosed, a high proportion (>66 percent) of them have reached an advanced, malignant, and aggressively metastatic stage that is not amenable to surgical intervention.1 Many patients with newly diagnosed lung cancer face a poor prognosis and a five-year survival rate of approximately 16 percent, according to the American Cancer Society (2012), despite the increasing range of systemic and targeted therapies that have become available.
The demand from clinicians and patients for clinically relevant real-time biomarkers that guide treatment decisions and prognosticate and monitor for response to therapy and/or disease progression, therefore, is high. The objective of this article is to review the newest developments in the field of biomarker testing for non-small cell lung cancer (NSCLC), and to demonstrate its emerging role as a standard of care in clinical management.
Biomarker testing in diagnosis and treatment
The mainstays of treatment for advanced NSCLC include generalized therapies (platinum-based or single-agent chemotherapies and radiation), immunotherapies (checkpoint inhibitors), second line without biomarker, Keytruda (checkpoint inhibitor) in frontline patients with high PD-L1 staining, and targeted tyrosine kinase inhibitor (TKI) drugs, which are directed at specific mutations in the epidermal growth-factor receptor (EGFR).2 However, several new developments are shaping changes in the NSCLC treatment landscape. For one, the number and breadth of targeted therapies for NSCLC continue to grow as new drugs are being brought to market. At the same time, it has become well understood that the presence of certain tumor biomarkers is highly predictive of an individual’s response to both generalized and targeted treatments, and this understanding is supported by new diagnostic tools capable of providing actionable information with which to make critical decisions about clinical management.
The profiles now provided by some commercially available biomarker tests can give physicians the opportunity to improve patient quality of life by reducing ineffective treatment. Such information can be used to customize the treatment plan, helping to avoid wasting precious time, causing debilitating therapeutic side effects, and dedicating financial resources to therapies that are not likely to work. Additionally, this information may suggest a benefit in pursuing subsequent broad molecular profiling which can identify rare mutations that might present options for therapeutic alternatives or clinical trials. Biomarker testing may help enable physicians to provide their patients with an objective, realistic assessment of prognosis and life expectancy to facilitate planning together for appropriate palliative care and end-of-life decisions.3,4
Basic research efforts into gene-expression profiling and genome-wide association studies continue to uncover potential correlations between the presence of genetic mutations and the aggressiveness of NSCLC and other cancers. A handful of these genetic aberrations—including point mutations, deletions, and fusions—have been proven to have direct predictive value with respect to treatment efficacy and prognosis in NSCLC. These so-called “driver” mutations sometimes perform a direct, mechanistic role in driving malignant transformation and cancer progression, and are often targets for current drug therapies. Some of these mutations include:
- EGFR sensitizing mutations (del19; (E746-A750); L858R): Mutations in the EGFR gene that render NSCLC cells sensitive to first- and second-generation EGFR-TKIs. Patients may benefit from treatment with afatinib, erlotinib, or gefitinib.
- EGFR resistance-conferring mutations (T790M): A second-site mutation associated with acquired resistance to gefitinib and erlotinib. Patients with this mutation may benefit from treatment with third-generation EGFR-TKIs osimertinib.
- ALK fusion variants (EML4 (E6:A20, E13-A20, E20-A20)): A translocation mutation that results in overexpression of the ALK kinase gene, resulting in dysregulation of critical downstream signaling pathways. Patients may benefit from treatment with crizotinib, ceritinib, or alectinib, depending on previous therapies.
- ROS1 fusion variants CD74 (C6:R34, C6:R32); SDC4 (S2:R32, S2:R34); SLC34A2 (S13del2046:R32, S13del2046:R34); EZR (E10:R34); TPM3 (T8:R35): Another genetic translocation that results in aberrant receptor tyrosine kinase activity. Patients may benefit from treatment with crizotinib.
- RET fusion variants KIF5B (K15:R12, K16:R12, K22:R12, K23:R12, K24:R11, K24:R8); CCDC6 (C1:R12); TRIM33 (T14:R12): Patients with mutations in this oncogene may benefit from treatment with cabozantinib or vandetinib.
- KRAS mutations (G12C, G12D, G12V): Mutations in this critical oncogene lead to constitutive activation of intracellular signaling cascades that normally regulate cell growth and differentiation. Prognosis is generally poorer in patients with these mutations.
- BRAF mutant V600E: Mutations in this serine/threonine-protein kinase adversely affect cell division and differentiation. Patients may benefit from vemurafenib, dabrafenib, or dabrafenib+trametinib.
Genetic and/or genomic testing panels that include these mutations (Table 1) are those that are most likely to provide actionable information to guide treatment strategies.
Table 1. Currently available genetic biomarker testing for NSCLC.
Turnaround time in biomarker testing
The clinical course of newly diagnosed, advanced NSCLC can be rapid, with prognoses typically measured in months. Early intervention with appropriately targeted therapies can increase the overall chance of survival, and therefore speed is critical when obtaining biomarker information that might influence the design and initiation of a therapeutic strategy. However, the results from tumor biopsies generally take three weeks or more, and inadequate sampling at the initial biopsy can lead to even longer wait times. According to a recent study, nearly 80 percent of patients diagnosed with NSCLC did not receive the results of biomarker testing in time for the initial consultation with their oncologist, and the median time to receive those results was 21 days after the initial consultation.5 When biomarker profiles were available at the initial consultation, patients experienced markedly shorter median times from consultation to decision (0 vs. 22 days, p = 0.0008), and to initiation of treatment (16 vs. 29 days, p = 0.004), compared to patients whose biomarker data were not yet available.
Timely availability of biomarker profiles is likely to improve due to the recent advent of so-called liquid biopsy for use in NSCLC biomarker testing.6 Liquid biopsy exploits the presence, in blood or other body fluids, of intact circulating tumor cells (CTCs) or cell-free, circulating tumor DNA (ctDNA) that is shed when tumor cells undergo apoptosis or necrosis. These cells and nucleic acids represent a readily available, easily accessible, and virtually limitless source of analytical material compared to tissue biopsy. In the last five years or so, the testing platforms that have been developed to analyze them have improved in sensitivity, speed, and cost to the point where they can achieve results much faster than techniques that begin with tissue as a starting material. With this technology, blood samples can be acquired on the same day as a tissue biopsy to yield rapid biomarker genotyping results.
Liquid biopsy is unlikely to replace the need for tissue biopsy in the near-term because information from tissue histology remains essential to diagnosing and subtyping the cancer.7 Liquid biopsy technologies are also limited in their ability to detect small, early-stage tumors that are not actively shedding material. However, due to its rapid time to results, biomarker testing of liquid biopsies represents such a powerful companion technique that the Cleveland Clinic included in its list of top ten medical innovations projected to impact patient care in 2017.8
Newer detection platforms
Liquid biopsy and other improvements in biomarker testing for NSCLC have been enabled by new diagnostic platforms that are faster and more sensitive than traditional immunohistochemistry, fluorescence in situ hybridization (FISH), and traditional rtPCR.
Droplet digital PCR (ddPCR) is a newly developed technique that uses water-oil emulsion droplet technology for highly sensitive, quantitative, reproducible detection of rare target mutations in ctDNA as well as cDNA copied from ctRNA.9–12 With this technique, processed nucleic acids from blood samples are diluted and partitioned into thousands of tiny droplets, each of which acts as an independent PCR amplification chamber. The results of amplification and detection yield absolute information regarding the quantity of mutations in the sample, independent of the amplification reference curves required for quantitative rtPCR experiments. Biomarker test platforms using ddPCR technology are very fast and highly sensitive (Table 1); median turnaround time for ddPCR testing in advanced stage NSCLC is three days, compared to 12 to 27 days for tissue genotyping assays.6 A recent study assessing performance of a ddPCR-based biomarker-testing platform demonstrated a sensitivity of 88 percent, specificity of 99 percent, and overall concordance with tissue biopsy of 96 percent. Thus, ddPCR is well suited for the sensitive analysis of small, defined panels of validated target biomarkers that yield clinically actionable information.
Next-generation sequencing (NGS) is a general term encompassing a variety of high-throughput, highly parallel DNA sequencing technologies that have replaced simple dye terminator sequencing methods.13 NGS enables relatively rapid, massively parallel analysis of large DNA segments or complex samples, including entire genomes. Originally developed to reduce the time and costs of genome sequencing, NGS technologies can analyze broad panels of genes for multiple mutations, all at the same time. In biomarker testing, NGS is well suited for liquid biopsy analysis and faster than more traditional methods, and it generally allows for analysis of larger gene panels than can be conducted with ddPCR (Table 1). However, much of the additional information currently yielded by NGS-based genomic panels is not actionable for NSCLC. Large panels have the potential to confuse clinical decision-making, because many of the genes and mutations are less well characterized, have not yet been validated, and are not yet part of recommended diagnostic guidelines.
Proteomic biomarkers provide complementary information
Another promising new direction for NSCLC biomarker testing is the availability of clinically validated tests that measure a patient’s response to a growing tumor using blood-derived proteomic information. Such tests take advantage of the complex biological interactions that occur as tumor cells begin to break free of normal growth control pathways, and the patient’s immune system responds with attempts to regulate and shut down this uncontrolled growth. This biological interplay between tumor and host releases a variety of acute-phase reactant proteins into the bloodstream. These can be detected and quantified, yielding important insights into the aggressiveness of the cancer and its susceptibility to available treatments.
So far, there is just one proteomic test that is commercially available for NSCLC. This proprietary test uses an advanced form of mass spectrometry, called matrix-assisted laser desorption/ionization-time of flight (MALDI-ToF), to detect and measure the chronic expression proteins associated with patient responses to aggressive cancer.14 The information yielded by this test can identify which patients are likely to benefit from first-line, standard-of-care platinum doublet therapy, and it is also predictive of therapeutic benefit from EGFR-TKIs.15,16 The results of the test have also been shown to be highly predictive of overall prognosis.14-16
Putting it all together: the reflex strategy
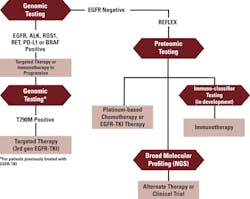
Tissue histology, genetic/genomic tests, and proteomic tests yield complementary information. Combining their results using what is termed a “reflex strategy” can be a powerful tool to help guide clinical management (Figure 1).
For example, a patient presenting with newly diagnosed NSCLC can undergo tissue and blood biopsy on the same day. Tissue histology confirms the diagnosis and aids in sub-typing the disease. Meanwhile, in as few as 72 hours after the blood draw, rapid genetic profiling from the liquid biopsy provides information regarding sensitizing EGFR mutations, as well as the presence of EML4-ALK, ROS1, or RET positive results, which indicate candidates for specific EGFR-TKIs and other targeted therapies.
For patients treated with EGFR-TKI therapy whose disease subsequently progresses, genetic profile testing can be performed again, without need for additional tissue biopsy, to detect EGFR mutations that confer resistance to EGFR-TKI, and to predict whether or not the tumor may be susceptible to third-generation EGFR-TKI inhibitors.
Patients whose initial genetic profile indicates the presence of wild-type EGFR, or whose EGFR status is unknown,15,16 can be reflexed to proteomic profiling to determine how aggressively their tumor is growing. A predicted prognosis of “good” indicates that a patient’s tumor is likely to respond to platinum-based therapy, single-agent chemotherapy, or EGFR-TKI-based therapies. A “poor” prognosis identifies aggressive cancer that is unlikely to respond to EGFR-TKI therapies and may also not respond to standard of care.
Availability of this information has been shown to change patient and physician decisions about treatment and supportive care in positive ways. They can preserve valuable time by shifting their focus from ineffective therapies to investigating broad genetic profiling, clinical trial options, or alternative treatments, thereby avoiding overtreatment that negatively impacts quality of life in terminally ill patients.
In conclusion, the adoption curve for biomarker testing for NSCLC is accelerating, driven by rapid improvements in analytical technologies, drug development, and evidence-based translational research. For clinicians, understanding the relevance and utility of the current state of biomarker testing is essential for optimizing outcomes and providing quality patient care. Laboratorians play a vital role in performing such testing and disseminating its results to oncologists.
REFERENCES
- Hurria A, Kris MG. Management of lung cancer in older adults. CA Cancer J Clin. 2003;53(6):325-341.
- National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology (NCCN guidelines): non-small cell lung cancer. NCCN website. https://www.nccn.org/professionals/physician_gls/PDF/nscl.pdf. Published 2017.
- Enzinger AC, Zhang B, Schrag D, Prigerson HG. Outcomes of prognostic disclosure: associations with prognostic understanding, distress, and relationship with physician among patients with advanced cancer. J Clin Oncol. 2015;33(32):3809-3816.
- Epstein AS, Prigerson HG, O’Reilly EM, Maciejewski PK. Discussions of life expectancy and changes in illness understanding in patients with advanced cancer. J Clin Oncol. 2016;34(20):2398-2403.
- Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. 2015;26(7):1415-1421.
- Bowling M, Mattingley J, Bhadra K, Pritchett M, Skibo S, Walker PR. PS01.16: Shortening time from diagnosis to treatment in NSCLC: are blood-based biopsies the answer? Topic: Pulmonology. J Thorac Oncol. 2016;11(11S):S278-S279.
- Mino-Kenudson M. Cons: Can liquid biopsy replace tissue biopsy?—the US experience. Transl Lung Cancer Res. 2016;5(4):424-427.
- Cleveland Clinic Newsroom. Cleveland Clinic unveils top 10 medical innovations most likely to be game changers. https://newsroom.clevelandclinic.org/2016/10/26/cleveland-clinic-unveils-top-10-medical-innovations-likley-game-changers/. Published 2016.
- Beck J, Bierau S, Balzer S et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 2013;59(12):1732-1741.
- Watanabe M, Kawaguchi T, Isa S, et al. Ultra-sensitive detection of the pretreatment egfr t790m mutation in non-small cell lung cancer patients with an egfr-activating mutation using droplet digital PCR. Clin Cancer Res. 2015;21(15):3552-3560.
- Whale AS, Devonshire AS, Karlin-Neumann G, et al. International interlaboratory digital pcr study demonstrating high reproducibility for the measurement of a rare sequence variant. Anal Chem. 2017;89(3):1724-1733.
- Zhang BO, Xu CW, Shao Y, et al. Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation. Exp Ther Med. 2015;9(4):1383-1388.
- Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11(1):31-46.
- Taguchi F, Solomon B, Gregorc V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99(11):838-846.
- Gregorc V, Novello S, Lazzari C, et al. Predictive value of a proteomic signature in patients with non-small-cell lung cancer treated with second-line erlotinib or chemotherapy (PROSE): a biomarker-stratified, randomised phase 3 trial. Lancet Oncol. 2014;15(7):713-721.
- Carbone DP, Ding K, Roder H, et al. Prognostic and predictive role of the VeriStrat plasma test in patients with advanced non-small-cell lung cancer treated with erlotinib or placebo in the NCIC Clinical Trials Group BR.21 trial. J Thorac Oncol. 2012;7(11):1653-1660.
Edward H. Kaplan, MD, is a medical oncologist in private practice in the Chicago area and an Assistant Professor of Medicine at Rush University in Chicago. He also serves as the medical director of the North Shore Cancer Research Association.