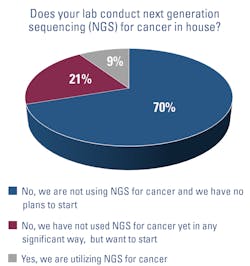
Hospital-based labs are beginning to deploy next-generation sequencing (NGS) for cancer diagnostics in-house, rather than sending samples out to specialized laboratories.
NGS allows labs to uncover the genetic makeup of tumors, helping oncologists determine if targeted therapies would be appropriate for their patients’ cancer. These therapies may be more effective and less toxic than standard chemotherapy and radiation.1
Despite the promise of targeted therapies, they do have limitations. Cancer cells can become resistant to the treatments over time, leading oncologists to use several therapies together or to pair them with standard treatments, such as chemotherapy.2
Nonetheless, the number of targeted therapies is continually increasing, making the case for bringing NGS in-house more compelling. For example, the U.S. Food and Drug Administration (FDA) approved 20 new therapies in 2020, according to an article in the May issue of Medical Laboratory Observer.1 Overall, there are hundreds of indications for targeted therapies, as the FDA has approved some medications for multiple types of cancer.2
Reducing turnaround and costs
Sentara Healthcare is one example. To improve test turnaround times (TAT) and decrease costs, the 12-hospital system decided to launch its in-house NGS service in 2017.
Before implementing NGS, Sentara did some sequential, single-gene analysis in-house, but samples often were also sent out to specialized labs for tests that the staff did not do.
It was a time-consuming process. “You could be looking at a week and up to a four-week turnaround to get a complete result back to a clinician. One of our major drivers was to be able to reduce that amount of time to result, and next-generation sequencing allows us to do that,” explains David Seidman, PhD, MB (ASCP)CM, Scientific Director of Molecular Diagnostics and Serology at Sentara Healthcare, based in Norfolk, VA.3
In-house NGS was not only a faster option for Sentara, but also a more cost“We are able to sequence a larger panel and provide a larger data set of results that also provides a physician with additional options for therapies and clinical trials that our patients can go on than we could obtain from just the single-gene analysis,” he says.
Currently, Sentara uses a commercial NGS panel for solid tumors. Most of the specimens that the lab sequences are non-small cell lung cancer, because there are therapies available that specifically target certain biomarkers in those tumors. “There are just more actionable targets for non-small cell versus other cancer types, so the panel contains genetic targets that would provide that actionable result for non-small cell,” Seidman explains.
Working with oncologists, the lab developed a reflex ordering process. If a pathologist determines that a biopsy is non-small cell lung cancer, he or she creates an order for NGS automatically and forwards the tissue block to molecular diagnostics. This saves time that would otherwise be used waiting for an oncologist to read the pathology report and then place the NGS order.
The goal is to provide patients with the results of the NGS test — as well as a treatment plan — at their first visit with an oncologist.
Although Sentara performs the bulk of in-house NGS testing on non-small cell lung cancer, it also sequences other solid tumors, such as for breast, colon, liver, and skin cancers.
The health system also would like to expand into other areas by adding new sequencing panels in the future, including for the analysis of tumor mutation burden and hematologic malignancies. Sentara currently sends out these types of tests to other labs.
Early NGS adopter
Meanwhile, Penn Medicine, an academic medical center based in Philadelphia, has been using NGS in cancer diagnostics since 2013 at its Center for Personalized Diagnostics, which runs NGS panels in hematologic malignancies, lymphoma, and solid tumors.4
At the time the center launched in 2013, it started with tests for solid tumors and hematologic malignancies, which it developed itself. “We were only sequencing AML (acute myeloid leukemia), brain tumors, lung tumors, and melanoma,” says Jennifer Morrissette, PhD, FACMG, Associate Professor and Laboratory Director for the CytoGenomics Laboratories at Penn.
“We decided to start with a small group of malignancies, because we weren’t really sure how well the clinical teams would receive genomic data for their patients.”
As it turns out, they found it to be clinically useful, and the program has grown substantially since then; it now sequences about 6,000 cases (not unique individuals) a year.
In 2020, the lab moved its panel for hematologic cancers to a hybrid capture-based technology. It plans to move its solid tumor panel to the technology this year.
Applying multiple methods
Sussman said hybrid capture-based technology allows the lab to sequence more gene targets than is possible with amplicon-based sequencing techniques. However, turnaround times for either method generally range from seven to 21 days, Morrissette added.
For patients with certain advanced cancers, that is too long to wait. Morrisette says most oncologists aim to have a rapid treatment plan or start treatment within days of a diagnosis. Oncologists want to know “whether to put them (patients) on a targeted therapy, immunotherapy or to go with the standard therapy,” she adds.
To solve this problem, the lab is in the process of evaluating a commercially available amplicon-based, end-to-end solution, which is faster than many other amplicon or hybrid capture technologies. It should turnaround results in 24 to 48 hours. While specialized training is still required, this NGS platform is relatively turnkey, so staff members who do not have a significant background in genomic assays can run the tests, Morrissette says. The lab plans to use this technology primarily for lung cancers.
The plan is to test the tumors with the quick method first — which focuses on genetic targets for which there are targeted therapies available — and follow up with more extensive testing afterward, if needed.
The lab also is evaluating whether to extend rapid testing to leukemia and other malignancies for which therapeutic decisions need to be considered in a matter of days post-diagnosis.
For all other cancers, Penn Medicine plans to stick with the larger panels.
In brain cancer, for example, the lab follows brain cancer patients with initial studies to provide information about the pathologic diagnosis. However, it also analyzes subsequent specimens, because the treatments can cause additional mutations, which could then become a target for a specialized treatment. In the case of gastrointestinal tumors, the sequencing studies will inform oncologists if certain treatments would be contraindicated. The tests also will detect targetable mutations.
Overall, Morrissette notes that cancer diagnostics is an exciting area of medicine to be involved in because it is rapidly evolving with new therapeutics, which can improve patients’ clinical outcomes. Detecting mutations that may enable physicians to treat some cancers as chronic diseases is the goal, she says. “Imagine if you can start extending the lives of more cancer patients by a few decades; that really matters.”
References
- Hampton G. In-sourcing next-generation sequencing supports patient care at community hospitals. Medical Laboratory Observer. May 2021. https://www.mlo-online.com/molecular/genomics/article/21218574/insourcing-nextgeneration-sequencing-supports-patient-care-at-community-hospitals. Accessed June 5, 2021
- Targeted cancer therapies. National Cancer Institute. https://www.cancer.gov/about-cancer/treatment/types/targeted-therapies/targeted-therapies-fact-sheet. Accessed June 8, 2021.
- Sentara cancer network–molecular testing laboratory. Sentara Healthcare. https://www.sentara.com/hampton-roads-virginia/medicalservices/services/cancer/clinical-trials-and-research/molecular-testing-laboratory.aspx. Accessed June 5, 2021.
- Penn Center for Personalized Diagnostics. Penn Medicine. https://www.pennmedicine.org/departments-and-centers/center-for-personalized-diagnostics/about-us. Accessed June 5, 2021.