From bench to bedside: the reality of the lab bench
The proper function of biological systems can be understood at multiple scales, ranging from the ecosystem and population level down to a molecular level, via individuals, organs, tissues, cells, and sub-cellular organelles. Here we are going to focus on the molecular level, well aware of the fact that studies of cause and consequence will have to take the connections to the other levels into account as well.
This narrow focus has the capacity to highlight underlying mechanisms of action as long as they do not become too complex, involving inputs from other levels. Indeed, the relative success of pharmaceutical research and drug development in recent years has built on mostly binary interactions between very specific biological molecules and drugs: simple hypotheses amenable to comprehensive testing in large patient cohorts. For more complex biological processes, such an approach becomes error-prone, and the popularity of systems biology has reflected this striving for a more effective approach.
Technology modifying the scientific approach
One enabling factor and driver of a systems approach in biology has been the increasing power of computers and an interest in the complexity of most biological processes and the fast-growing amount of raw measurement data from people with training in theoretical physics and mathematics. Thus, for the first time, biology has come into the center of high-power computing and the world of databases.
This evolution was stimulated by the fast development of high-throughput measurement technologies such as microarrays and sequencers, in particular sequencers, first as a consequence of global projects on the model species yeast and fruit fly, but then also the human genome and other genome projects. The first human genome was delivered in 2003. The microarray and sequencing technologies, complemented by the development of mass spectrometry toward higher throughput, changed the orientation of the scientific approach toward the quantitative analysis of a complete or near-complete assortment of all the constituents of a system. The term “omics” entered the English language: genomics, transcriptomics, proteomics, metabolomics, etc.
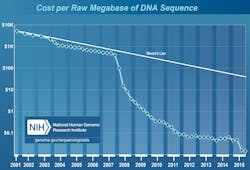
Genomics is the study of whole genomes, or at least of more than one gene at a time, out of the approximately 20,000 genes in the case of the human. The arrival of next generation sequencing (NGS) allowed genomics to spread outside the confines of research laboratories as cost dropped faster than it had with regard to other technologies (Figure 1). NGS, like the older microarray technology, is about probing the sequence of nucleic acids—in other words, the reading of the code of genes in the genome. In essence, NGS is just sequencing as it has been for half a century. The biggest difference lies in the efficiency, the cost, and thus in the growing availability of the technology.
The NGS measurement concept has some important peculiarities. First, the measurement is an average of many values, not one single measurement. Therefore, the interpretation of NGS data requires a radically different computational approach. Many computational algorithms have been developed to cope with different aspects of the statistics and error handling of the data processing pipeline. There is today no single solution universally applicable for NGS data processing that would yield both high precision and high sensitivity. The technology has to operate with concepts such as false positives and false negatives, in addition to true positives and negatives. Optimizing between these parameters requires understanding of the sequencing technology biases and computational processes and availability of reference material.
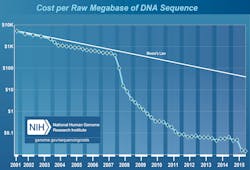
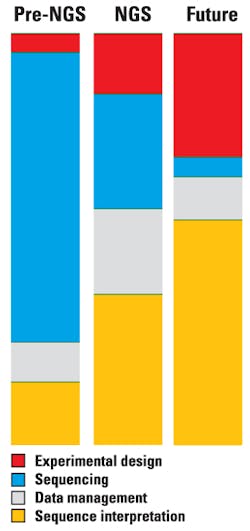
This leads us to the second peculiarity. As the cost of sequencing continues to decrease and the amount of sequence data generated grows, the cost of the computational part of sequencing grows. The real cost of sequencing (Figure 2) is far higher than the often cited numbers, such as the $1,000 genome (as shown in Figure 1). New paradigms for data storage and analysis are increasingly important, and the relative scaling behavior of these evolving technologies will impact genomics both in research and in the clinic.1
With NGS, the clinical diagnostics world is opening to new opportunities: diseases with a genetic component become amenable to much more precise diagnostics than before, at a price worth considering in healthcare. NGS is now sparking a profound change in the world of diagnostic molecular genetics.
High-throughput genetics in medicine: the permanent and the changing
The study of nucleic acids encompasses not only the DNA of the genome, which is the constant part of any organism. All cells of an individual (with some exceptions) carry the same genome, which basically remains constant over the span of a lifetime. Every individual has a unique genome that carries information that will affect most aspects of that individual’s life. However, there is also a variable component among nucleic acids. All cells are not identical, and cells need to respond to changes in the environment or over time. Much of this capacity for variability is provided by the changing use of the constant genome—in other words, by changes in gene activity. Far from all genes are constantly active, despite being ubiquitously present in the genome. The variations in gene expression, that is, in timing or quantity, provide important information about the function of the cell and can be measured on the level of RNA molecules, a product of gene activity. Changes in both DNA and RNA can be connected to health, risk, or disease, and both can be analyzed with NGS.
DNA and RNA sequencing have until recently been confined to research center core facilities with sufficient budgets to buy sequencers in the million-dollar range that need an upgrade as often as a smartphone. Today, a desktop sequencer that can handle a tenth of the human genes for a dozen samples in one run—which is substantial from a typical diagnostic perspective—can be purchased for the price of a six person-month laboratory specialist salary. Therefore, NGS technology is now found in an exponentially growing number of labs, disrupting several aspects of diagnostics, not only in human healthcare but also in food testing, cattle breeding, etc. The rise of NGS has fuelled invaluable large projects, such as the Cancer Genome Atlas, the 1,000 Genomes Project, and the 100k Genomics England project.
The complexity of biological processes remains a major issue in medicine. This can be translated into a problem of diagnostics: which genes and which parts of the genome to measure for each different medical case. The knowledge of the biological processes involved in health and disease is often fragmentary and incomplete, but it is also progressing rapidly. What one would consider today as relevant genes to probe for a particular disease can change quite dramatically in the course of a year of clinical research. Harnessing the predictive power of genomic knowledge is even more elusive, although some business is already being done based on the buzz.
Despite the complexity of biological systems, the search for and study of simple relationships continues to yield clinically actionable data, but large areas still remain uncovered. Today, much of the “simple” has been found and turned into medical understanding or drugs. Hence we face the “less simple.” Complex diseases such as diabetes, neurological disorders, cardiovascular disorders, and cancer are only partly understood, and the genetic component is only one player alongside nutrition, life- style, and others. Much of the buzz around genomics is still sheer dreaming, but it helps to stimulate further innovation and, by the way—some dreams have already come true!
NGS in diagnostics today: a few examples
Molecular genetics diagnostics based on NGS evolves fast. There is a strong mutual stimulation between the bench and the bedside in the use of NGS-derived analytics. For example, acute myeloid leukemia (AML) is defined, classed, and staged based on genetic, genomic, and molecular characteristics.2 The use of certain drugs can be dictated by the presence or absence of specific changes in the genome. This is obvious if one considers that many drugs will target proteins, the 3D structure of which is on one hand critical for the interaction with the drug, but on the other hand also coded by the corresponding gene sequence. A mutation can change the 3D structure or chemical properties of the docking site of a drug, thereby modifying the effect of the drug.
Other mutations can change the metabolic processing of drugs. The dosage of the anticoagulant warfarin is problematic since there are large individual as well as inter-population variations in its metabolism. Single nucleotide polymorphisms (SNP) in genes such as CYP2C9 and VKORC1 affect warfarin dosage tolerance because these genes affect the metabolism of warfarin.3 Pharmacogenomics is an area of genomics concerned with the effect of genetic variations on the absorption, distribution, metabolism, and excretion of drugs.
NGS-based molecular genetics can also replace other forms of diagnostics, when looking at the overall cost of diagnosis. A recent prospective evaluation of NGS suggests that whole-exome sequencing could be a first-tier molecular test in infants with suspected monogenic disorders, replacing more traditional and less efficient methodologies.4
From bench to bedside: the reality of the bedside
Western medicine is mostly evidence-based. Evidence gathered from the study of simple biological relationships between molecule and function has yielded a considerable amount of new drugs and treatments to the benefit of patients. However, with the avenue of high-throughput technologies, the concept of evidence has changed. It has become more dependent on statistical significance and on sophisticated data mining methods—the interdisciplinary activity of discovering patterns in large data sets involving methods at the intersection of artificial intelligence, machine learning, statistics, and database systems. Here, evidence is more elusive, and the adoption of high-throughput technologies at the bedside poses new challenges. Diagnostics based on these technologies does not develop as fast as the technologies themselves. Genome or transcriptome measurements are not often affecting medical treatment.
Precision medicine acknowledges that patients are different and require an individually adapted treatment. Yet evidence for clinical applicability has so far come from the use of large test cohorts. These two concepts are somewhat in conflict, and healthcare will have to revise its requirements for evidence from large cohorts. However, precision medicine itself should help in this process, as it implies more targeted effects and fewer side effects of treatments, and thus evidence from much smaller cohorts.
Precision medicine is also related to companion diagnostics, where a drug is developed in conjunction with a specific diagnostic test that shall provide an indication of the suitability of the drug. Many companion diagnostic tests are array- or NGS-based molecular genetics tests.
Diagnostics and treatment tomorrow
For genome reading technology, be it microarray-based or now mostly NGS-based, to be able to provide diagnostic data that can be used with confidence in healthcare, much still needs to be done. The quality of the data and standards applied to monitoring this quality are still insufficiently talked about, but are of pivotal importance. This aspect is being addressed by proficiency tests developed by various accreditation bodies and by best practice guidelines developed by organizations such as the College of American Pathologists, EuroGentest, etc.
While diagnostics is an important part of the medical process, other aspects based on knowledge of genome sequence and function are emerging. Most notably, gene editing makes it possible to take the step from diagnosis to treatment. Advances in the development of genome editing technologies based on programmable nucleases (nucleic acid-cutting enzymes, e.g. CRISPR-Cas9) have substantially improved the ability to make precise changes in the genomes of cells.5 A particularly tantalizing application of gene editing is the potential to directly correct genetic mutations in affected tissues and cells to treat diseases that are refractory to traditional therapies. The ethics of human-genome editing will be an important matter of debate far beyond the much less problematic yet fundamentally related topic of genome reading.
REFERENCES
- Muir P, Li S, Lou S, et al. The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biology 2016;17:53-61.
- Forthun RB, Hinrichs C, Dowling TH, Bruserud Ø, Selheim F. The past, present and future subclassification of patients with acute myeloid leukemia. Curr. Pharm. Biotechnol. 2016;17(1):6-19.
- Yang L, Ge W, Yu F, Zhu H. Impact of VKORC1 gene polymorphism on interindividual and interethnic warfarin dosage requirement—a systematic review and meta analysis. Thromb. Res. 2010;125(4):e159-66.
- Sawyer SL, Hartley T, Dyment DA, et al. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin. Genet. 2016;89(3):275-284.
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nature Medicine 2015;21/2:121-131.
- Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB. The real cost of sequencing: higher than you think! Genome Biol. 2011;12(8):125-134.
Christophe Roos, PhD, is assistant professor in developmental genetics at the University of Helsinki and serves as CSO for Espoo, Finland-based Euformatics, provider of omnomicsQ and omnomicsNGS for genomic data quality control and variant interpretation.