Genetic analysis has come a long way; we now have an ever-expanding collection of analytical tools in the diagnostic laboratory. So why do we still need a technique that usually only looks at one or two loci? The simple answer is that results from fluorescence in-situ hybridization (FISH) can quickly confirm diagnoses, guide clinicians’ judgements regarding differential diagnoses, and correlate results with clinical risk—thus enabling an informed choice of treatment type and intensity.
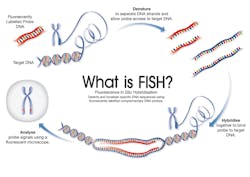
FISH employs fluorescently-labeled DNA probes to bind complementary DNA sequences within an interphase cell, or onto metaphase chromosomes. These sequences can then be visualized using fluorescent microscopy. The number, location and relative positions of the probe signals indicate chromosomal changes in a particular cell (Figure 1).
Many clinical trials use cytogenetic and FISH data to stratify patients according to specific risk factors. FISH is often used as a stand-alone technique for investigating abnormalities and following-up such patients, which, alongside its relatively low expense, makes it a very convenient investigative tool.
In this article we will explore the utility of FISH in today’s clinical laboratory and the future of the technique in the evolution of molecular testing.
A popular technique
FISH is a technique that has been in common use in the laboratory since the 1980s. Part of the reason for its ongoing popularity is that it is easy to set up. The visual nature of the technique gives an immediate indication of whether the process has worked, and FISH testing validation is relatively straightforward.1 The downside of this visual nature is that signal patterns may be ambiguous or open to interpretation due to the type and heterogeneity of biological material. There are a number of software-based FISH systems available that can scan, assess, and categorize large numbers of cells automatically; however, the price of such systems may be prohibitively high, and the software does not resolve potential signal pattern ambiguity.
In contrast to G-band cytogenetic analysis—an accepted technique for almost 50 years—the utility of FISH is not confined to dividing cells; FISH techniques, like molecular testing, can be applied to formalin-fixed paraffin-embedded (FFPE) specimens.
FFPE FISH is a potent adjunct to immunohistochemistry (IHC) for histopathology samples; it allows the analyst to visualize abnormalities in the context of specimen architecture. Using an H&E stained slide, the analyst can be guided to areas of interest. FFPE FISH can therefore be used to determine focal areas of genetic change and can assess tumor heterogeneity, both of which are hard to determine using molecular techniques alone.
FFPE FISH presents its own problems, however, as the process of slicing FFPE sections truncates cells. This means that a proportion of FISH signals in an FFPE specimen will be lost—a serious confounder when using FISH to assess the presence of a deletion in a specimen.
Clinical utility
Clinically, FISH is performed in both oncology and constitutional (cytogenetic) settings. In oncology, the detection of recurrent acquired abnormalities is the foundation of diagnostic pathways, risk stratification, and therapeutic decisions. In a constitutional laboratory, FISH screening may be carried out pre- or post-natally to identify a wide range of syndromes or conditions.
The vast majority of oncologic FISH investigations are carried out on bone marrow samples for the investigation of hematological or lymphoid malignancy. Often, patients with acute leukemia will present with a high percentage of abnormal cells in their bone marrow; however, in many cases the number of abnormal cells within the testing sample may be low. Targeted FISH can reliably demonstrate the presence of a gene fusion in as few as one percent of cells when using a dual-fusion probe set2 and can also, in some cases, demonstrate clonal evolution, progression events, or the presence of multiple cell lines. Two important applications of FISH are seen in the diagnosis of patients with chronic myeloid leukemia (CML) and acute promyelocytic leukemia (APL).
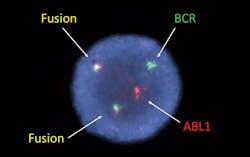
The majority of patients with CML harbor a characteristic gene fusion: the BCR-ABL1 rearrangement3 (Figure 2). FISH analysis for the BCR and ABL1 regions can detect almost all variants of this rearrangement. FISH is performed concurrently with quantitative PCR, and the results are interpreted in conjunction. Although FISH testing sensitivity is much lower than quantitative PCR, it can provide important additional information with regard to potential blast transformation if additional abnormalities are present.
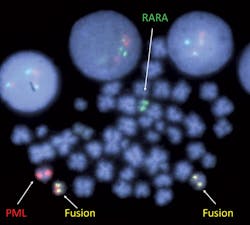
The diagnosis of patients with APL hinges on detection of a pathognomonic gene fusion, PML-RARA4 (Figure 3). Early diagnosis of patients with APL is important as, without treatment, this disease can cause serious clotting or bleeding problems.5 Thus, rapid testing is required in situations where APL is suspected. Recent advances in FISH probe formulation have enabled the manufacture of FISH probes that can be hybridized in just a few hours, meaning that a PML-RARA FISH result can be reported to the clinician on the same day, something that, at present, sequencing is unable to do.
FISH is also particularly important in the diagnosis of acute lymphoblastic leukemia (ALL), where a number of FISH-targetable recurrent abnormalities have been demonstrated to be important in risk stratification.6 In other hematological malignancies, for example chronic lymphoid leukemia (CLL) or multiple myeloma, difficulty in obtaining conventional cytogenetic G-banded preparations has caused an almost complete shift to FISH testing for these diseases. This has resulted in FISH-based risk stratifications and, in some cases, the use of companion diagnostics.7-8 In CLL, copy number changes are the major FISH assessments, but lately there has been a shift away from FISH testing toward array-based molecular testing. This is probably because the cost of a single array has become low enough to compete with the cost of FISH tests for CLL investigations. Array analysis also has the additional benefit of providing information about many more loci than the four single FISH tests.
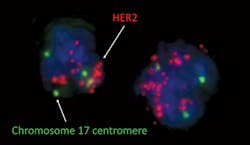
Although FISH testing is carried out on many types of solid tumor specimens, the majority of clinically relevant testing on solid tumors is carried out using probes for HER2 and ALK on breast cancer and non-small cell lung cancer specimens, respectively. In breast cancer, FISH testing confirms HER2 amplification status by counting the number of copies of the HER2 locus (Figure 4). It can demonstrate small areas of amplification, which may be equivocal by IHC testing, or be missed by molecular testing.
(All images courtesy of Oxford Gene Technology)
ALK FISH testing is carried out using a break-apart probe which will indicate the presence of ALK gene rearrangement. Cases that are equivocal by IHC testing may show small areas of ALK rearrangement-positive cells by FISH.
FISH testing is also still carried out in constitutional laboratories, primarily for rapid prenatal testing as well as post-natal testing for microdeletion syndromes, although the post-natal use of FISH has declined with the uptake of array testing. These molecular testing methods have a higher resolution than FISH and also target many loci simultaneously, giving a more comprehensive result. FISH may, however, still be used in these laboratories to confirm array results.
FISHing in the future
So, what does the future hold for FISH? Sequencing methods have historically handled the role of mutation analysis due to FISH’s inability to detect these aberrations, while FISH testing has been used to identify copy number neutral changes such as translocations, inversions, or insertions in the diagnostic laboratory. Recently, however, next-generation sequencing (NGS) methods have become capable of predicting copy number neutral changes and, as more molecular testing is carried out at diagnosis, further risk stratification will occur based on that testing. At present, these analyses are assessed alongside cytogenetic and FISH testing results,9 but based on recent trends, the time will come when molecular stratification will be the sole assessment criteria.
Will this spell the demise of FISH? Quite possibly at some point, but the technique will be around for a long time yet. As the cost of NGS decreases, more labs may switch to NGS; however, conventional cytogenetics is still the cheapest and easiest whole genome screening test—albeit at very low resolution—and FISH testing dovetails nicely into a cytogenetics lab set-up.
FISH is particularly useful in FFPE analysis, as molecular methods can be hampered by poor input DNA, cannot show focal areas of abnormality, and cannot be correlated with architecture. FISH can reveal tumor heterogeneity, which may be missed by conventional molecular analysis.
The shift towards molecular testing is perhaps inevitable, but as an inexpensive, robust, and reliable test that is easy to implement, FISH testing will be around for a long time to come.
Visit www.mlo-online.com for references.
REFERENCES
- Wiktor AE, Van Dyke DL, Stupca PJ, et al. Preclinical validation of fluorescence in situ hybridization assays for clinical practice. Genet Med. 2006;8(1):16-23.
- Shi M, Cipollini MJ, Crowley-Bish PA, Higgins AW, Yu H, Miron PM. Improved detection rate of cytogenetic abnormalities in chronic lymphocytic leukemia and other mature B-cell neoplasms with use of CpG-oligonucleotide DSP30 and Interleukin 2 stimulation. Am J Clin Pathol. 2013;139(5):662-669.
- De Braekeleer E, Douet-Guilbert N, Rowe D, et al. ABL1 fusion genes in hematological malignancies: A review. Eur J Haematol. 2011;86(5):361-371.
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues, Fourth Edition. 4th ed. IARC Press, Lyon, France; 2008.
- Sanz MA, Grimwade D, Tallman MS, et al. Management of acute promyelocytic leukemia : recommendations from an expert panel on behalf of the European LeukemiaNet Review article Management of acute promyelocytic leukemia : recommendations from an expert panel on behalf of the European LeukemiaNet. 2009;113(9):1875-1891.
- Moorman A V., Harrison CJ, Buck GAN, et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109(8):3189-3197.
- Fonseca R, Bergsagel PL, Drach J, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210-2221.
- Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910-1916.
- Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2015;33(0).
Steve Chatters DipRCPath, serves as Director of Medical Affairs for Oxford Gene Technology (OGT). Before joining OGT, he worked as a Principal Clinical Scientist, with over 20 years’ experience in specialist cancer diagnostic laboratories. He continues to act as a solid tumor cytogenetics examiner for the international Cytogenomic External Quality Assessment Service (CEQAS), and is a member of the CEQAS Specialist Advisory Group for tumor cytogenetics.