The clinical value of next-generation sequencing integration within medical laboratories
Sanger sequencing commonly utilizes clonal amplification of adaptor-ligated DNA fragments across the surface of a glass flow cell, yet it is limited in terms of low throughput and complexity.6 Major improvements and advancements in molecular biology that were transitively incorporated into sequencing technologies led to the development of second and third generation sequencing methodologies, commonly termed next-generation sequencing (NGS). Such innovations led to the milestone achievement of completion of the human genome project.1,3,6 Nowadays, sequencing turnaround time and cost have dramatically reduced, as well as have become more automated and compact since the early 2000s, thus enabling easier adoption and more practical widespread utilization within the clinician setting and beyond.2,3
An immense amount of curated clinical, genetic, and genomic data has emerged through NGS, helping foster the development of more precision based medicine, laboratory diagnostics, and clinical treatment.2,3 In addition to microorganism identification, NGS has been utilized for detection of antibiotic resistance, single nucleotide polymorphism (SNPs), and the host immune response.3 The clinical value of NGS has been exemplified, not only at the individual patient level, but as well as, NGS has been utilized to help govern and direct public health and (hospital) infection control strategies. For example, NGS contributed (and still contributes) to the discovery and tracking of SARS-COV2 variants including alpha, delta, omicron, and possible future variants throughout the course of the ongoing global pandemic and currently, Public Health England routinely employs whole-genome NGS to track spread of antimicrobial resistance of M. tuberculosis.7,8
Given these developments, the U.S. Food and Drug Administration (FDA) has outlined the guidelines for designing, developing, and validation of approved NGS tests.3 Generally speaking, both second and third generation sequencing technologies share nearly identical three step workflows: (1) preparation and extraction of nucleic acid template; (2) preparation of library including clonal amplification; and (3) sequencing and alignment of short reads.3
Science and methodology of NGS within the laboratory:
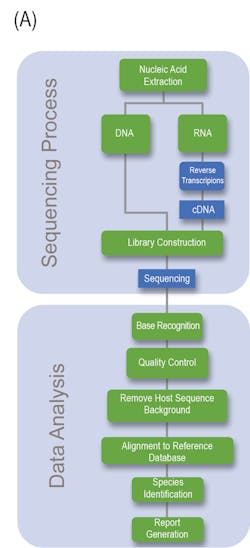
Generally speaking, NGS can be divided into the Sequencing and Data analysis phase (Fig 1A). With regards to the clinical lab, NGS possesses several steps and variables that must be taken under considerations if a clinician or laboratory manager desires to implement NGS within its clinician pipeline, the details of which are outlined in Fig 1B.
Nucleic Acid Extraction: Similar to nearly all NAAT based assays, the first step is nucleic acid extraction from the specimen. Due to the enhanced sensitivity and capability of detecting DNA or RNA from any organism, precaution needs to be taken to limit the risk of contamination of the extraction reagents. For example, the commensal flora of laboratory personnel can contaminate laboratory reagents and risk leading to inappropriate patient diagnoses.5
Library Preparation and Clonal Amplification: Post extraction of sample nucleic acid (whether it be DNA or RNA), the specimen is further processed in order to ensure compatibility and optimization for high-throughput sequence analysis. Library preparation is a delicate process comprising several steps that seek to preserve or enrich the pathogen sequences present within the sample, while maintaining the complex, native diversity, that is intrinsic to the sample. Depending on the type and target of the NGS assay (targeted, whole genome, or metagenomic, discussed later), pathogen genetic material can be selectively enriched using differential lysis, DNase or RNAse, mitochondrial and/or ribosomal RNA depletion, or whole genome hybridization. However, most clinical laboratories will likely employ an unbiased strategy utilizing total nucleic acid to more broadly identify for the presence of pathogen DNA. If a more targeted or refined approach is desirable, commonly, spiked targeted primers specific to conserved regions of either bacteria (16S rRNA), fungal (internal transcribed spacer region) or to different clades of viral targets will be added.3,5,10
The final step required for creation of the library is the addition of sample barcodes and sequencing adaptors, using standard, common techniques. Sample barcodes are short DNA sequences ligated to the ends of each sample library that allows for the pooling of multiple samples for sequencing analysis and sample identity using bioinformatics. Sequencing adaptors are specialized, and specific oligonucleotide adaptors tailored to a given sequencing platform and are commonly added through either adapter ligation or transposase-mediated addition.5
Sequencing: Over the past decade, a massive amount of commercially available sequencing platforms has emerged that offer high-throughput analysis. To generate sufficient data for adequate sequencing analysis, most platforms will pool libraries for sequencing. Quantification of the pooled libraries can be employed using several approaches, such as total DNA quantification, quantitative PCR normalization, and bead normalization. Within the clinical setting, several factors need to be taken into consideration when performing or considering NGS sequencing. All sequencing platforms have an intrinsic error rate that needs to be considered for data analysis. Further considerations include the level of throughput for the number of total sequences obtained as well as their profile length; the number of base pairs obtained; the sequencing depth per sample; and the physical computational hardware for processing and storing large NGS data files.5
The Generations of Sequencing Platforms: Post the advent of first generation of sequencing technology of Sanger Sequencing, second and third generation have emerged has the technology has advanced. The umbrella term of NGS includes second and third generation sequencing. Second generation requires template amplification prior to sequencing, while third generation offer de novo assembly in real time without the need of template amplification.6
Platforms such as Ion Torrent, Pacific Biosciences, and Illumina are the current frontrunners of second generation sequencing technology.3 Ion Torrent is unique in its detection method. Unlike other technologies that use fluorescence or chemiluminescence, Ion Torrent detects proton release during nucleotide incorporation of strand synthesis.3
Second generation sequencing has significantly revolutionized and advanced the field, yet the technology is not without flaws. Second generation sequencing typically has short sequence reads leading to sequencing gaps, alignment issues due to repetitive regions/pseudogenes, and PCR artifacts.3 As a means to overcome these limitations, third generation sequencing, offering sequencing at the signal molecule level, was developed. PacBio SMRT and Oxford Nanopore Technologies are the current representatives of third-generation sequencing.3,6
PacBio SMRT has a similar library preparation except for specialized adapters to circularize double-stranded DNA fragments. The circularized DNA and DNA polymerase are immobilized and analyzed on a chip. The signal from the incorporation of fluorescently labeled nucleotides is measured via a CCD camera.3
Oxford Nanopore uses a novel technology called nanopores. Nanopores are tiny bio-pores with nanoscale diameter, capable of measuring current changes. Each of the 4 types of nucleotides will pass through the nanopore, altering the channel voltage, and lead to a distinct current change that is measured by the platform. Nanopore technology is advantageous of short turnaround time and no GC bias, yet has the disadvantage of nanopore technology is its high sequencing error rate.3
Bioinformatic Data Analysis: Bioinformatic sequencing data analysis involves a multistep well-established pipeline as a means to identify any pathogen sequences present in the sample. Generally speaking, the sequential major steps are quality filtering, human subtraction, alignment to a (pathogen) database, taxonomic characterization, and genome mapping. The confidence of the sample is proportional to the number of sequence reads identified for the organism, normalized to the total number of reads present within the sample, and the overall genome coverage. Optional quantitative controls enable for the determination of the number of molecules per milliliter of organism DNA in the original sample to be determined.5 The direct clinical application of NGS to detect infectious agents is contingent on the availability of a curated databases to provide a high level of confidence of matched reads against the organisms identified. For example, organism types may not be present in the database thus hampering their detection. Though nucleotide alignment is the most commonly employed analysis strategy, amino acid /protein can be utilized to identify possible divergent organisms.5 NCBI possess a vast amount of curated and uncurated databases that are ever expanding. For example, more than 376,000 bacterial genomes are currently available.5,6
In order to offset the magnitude of data achieved from NGS a number of software platforms have been developed, a significant investment cost that could likely hinder more universal acceptance within the clinical lab.11 A number of software platforms, both commercially available (bioMérieux Episeq, Illumina, Bio-Rad’s SeqSense, Qiagen’s OmicSoft Suite) and open source software suites, are readily available. Episeq, designed developed by bioMérieux; as well as, several open source platforms provide cloud-based computing thus offering an attractive alternative to limited in-house analysis.11
Clinical utility and interpretation of the report analysis of NGS:
Similar to all diagnostic assays, the clinical, real-world utility of NGS testing is dependent on a number of critical factors to consider: (1) the patient presentation, symptom severity, and timing of sample collection; (2) the sample quality, location, and infection source type; and (3) the native operational characteristics of the assay, including but not limited to analytical sensitivity, specificity, and detection range.5
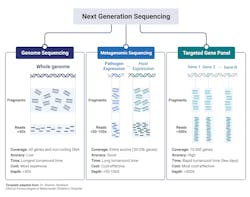
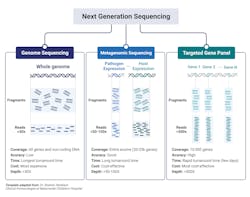
The major clinical application within microbiology laboratories are: whole genome sequencing, metagenomic NGS (mNGS), and targeted NGS (tNGS) (Fig 2).3,10 Whole-genome sequencing involves sequencing and assembly of an entire template within a clinical sample, enabling simultaneous typing of any microorganism or virus genome; and in some cases, identifying resistance gene/mutations/prediction of antimicrobial susceptibility of a given strain.2 Generally speaking, a pure sample of colony is needed for this approach.8 A more mass and widespread approach, metagenomic sequencing, involves sequencing all available templates within a clinical sample including pathogen and human DNA and RNA. This approach is advantageous in that it does not requiring culturing and takes an unbiased approach, enabling for the detection of numerous pathogens (and the associated host response against them). Finally, targeted NGS is similar to metagenomic yet is more refined, focusing on a subset of genes. Targeted NGS first enriches for sequences of interest before the preparation to enhance analytical sensitivity. Contingent upon the focus of the specific disease or disorder, this can range from several to a few hundreds of gene targets.3,10 For example, panels can be specialized to target bacteria, viruses, and eukaryotic pathogens.3
Whole genome sequencing’s ability to sequence and assemble an entire genome with plasmids is advantageous within the clinical laboratory as a means to identify antimicrobial resistance profiles thereby influencing first-line drug implementations. Recently, whole-genome sequencing proved beneficial to detect and characterize the emergence of several patients suffering from pneumonia from an unknown cause from Wuhan, China. While the initial sequence was of unknown origin, later bioinformatic analysis identified similarity to beta-coronaviruses and termed SARS-CoV-2 thus exemplifying the benefit of whole genome sequencing in identifying novel organisms and/or mutations.3,10
Metagenomic NGS has proven beneficial when targeted or less comprehensive tests fail. Given its wide inclusivity and lack of requirement of previous knowledge of potential pathogens, several clinical tests have been developed from a variety of patient samples including: synovial fluid, CSF, feces, corneal tissue, blood, plasma, nasopharyngeal swabs, and joint fluid as a proxy to diagnose various types of infections.3,10 Body fluid samples can possess significant complexity in terms of the biodiversity present and metagenomic NGS enables for detection of low-prevalence templates within the entire sample that would have likely been missed by other diagnostic means.3
A limitation of Metagenomic NGS is the disproportionate ratio of host to pathogen nucleic acid reads thus decreasing the analytical sensitivity of the assay. Targeted NGS improves analytical sensitivity by first enriching for highly conserved regions of pathogens, such as the 16S rRNA in bacteria. Targeted NGS has proven beneficial in terms of contributing to public health, such as enriching for SARS-CoV-2 RNA in clinical samples as a means to track the rise of variants.3,10 Targeting NGS of both the host and its associated flor3a can serve as an indicator of the general well-being of the patient. For example, sequencing of the gene expression of a patient’s immune response gene profile combined with sequencing of commensals and pathogen genomes lead to the correct identification of the causative agent with high sensitivity and specificity with a true negative predictive value of 100%. Likewise, sequencing of the virome within immunocompromised patients can serve to evaluate the competency of the host immune system, if viral loads dramatically increase under immunosuppressants.
These examples highlight just a few examples of the massive degree of publications available of NGS. It is wide-accepted that NGS possess immense value in contributing to the clinical utility within the healthcare setting. However, these assays are not without flaws. Contrarily, advances are ever ongoing to help contribute to easier adaption within the clinical lab and better patient outcomes.3
The Current Limitations to Widespread Implementation: It is accurate to claim that NGS sequencing as a diagnostic tool is still in its infancy.10 Currently, the most commonly utilized NGS platforms are limited by short reads, reliant upon clonal PCR, have high error rate, requires advanced technical expertise, and guidelines are not universally standardized.3,10 The process of implementing NGS sequencing requires significant resource investment, including test validation, bioinformatics support, data storage, and overcoming insurance cost hurdles.3 Most testing is current limited to reference laboratories or academic research centers that can afford such upfront resource investment.2,10 It is reasonable to assume that as the technology continues to improve to and advance, the threshold for more widespread adaption will decline.10 As with any molecular based diagnostic assay, testing results alone do not guarantee infection and the asymptomatic colonization.4
Conclusion:
Within clinical practice NGS possess mass potential, but as it stands today the most optimal, practical utilization appears to be in patient populations where infection is strongly suspected, yet conventional testing is negative.10 The field would significantly benefit from a prospective, controlled clinical trial evaluating the clinical utility for unbiased pathogen detection from clinical samples. As it stands today, the majority of publications comprise case reports and retrospective studies comparing the results to the traditional standard of care.10 It is likely only a matter of time before completion of such types of studies; thus, such research articles would allow for a more convincing argument for clinicians to adapt the application more readily. Likewise, as continual refinements and improvements to the technology continually emerge, NGS can and will be more easily integrated and streamlined within the clinical setting.
References:
- Duan H, Li X, Mei A, et al. The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases. BMC Infectious Diseases. 2021;21(1)doi:10.1186/s12879-020-05746-5
- Patel R. Advances in Testing for Infectious Diseases-Looking Back and Projecting Forward. Clin Chem. Dec 30 2021;68(1):10-15. doi:10.1093/clinchem/hvab110
- Zhong Y, Xu F, Wu J, Schubert J, Li MM. Application of Next Generation Sequencing in Laboratory Medicine. Annals of Laboratory Medicine. 2021;41(1):25-43. doi:10.3343/alm.2021.41.1.25
- Curren EJ, Lutgring JD, Kabbani S, et al. Advancing Diagnostic Stewardship for Healthcare-Associated Infections, Antibiotic Resistance, and Sepsis. Clinical Infectious Diseases. 2022;74(4):723-728. doi:10.1093/cid/ciab672
- Miller S, Chiu C. The Role of Metagenomics and Next-Generation Sequencing in Infectious Disease Diagnosis. Clin Chem. Dec 30 2021;68(1):115-124. doi:10.1093/clinchem/hvab173
- Ben Khedher M, Ghedira K, Rolain J-M, Ruimy R, Croce O. Application and Challenge of 3rd Generation Sequencing for Clinical Bacterial Studies. International Journal of Molecular Sciences. 2022;23(3):1395. doi:10.3390/ijms23031395
- Eyre DW. Infection prevention and control insights from a decade of pathogen whole-genome sequencing. Journal of Hospital Infection. 2022;122:180-186. doi:10.1016/j.jhin.2022.01.024
- Datar R, Orenga S, Pogorelcnik R, Rochas O, Simner PJ, van Belkum A. Recent Advances in Rapid Antimicrobial Susceptibility Testing. Clin Chem. Dec 30 2021;68(1):91-98. doi:10.1093/clinchem/hvab207
- Parker K, Forman J, Bonheyo G, et al. End-User Perspectives on Using Quantitative Real-Time PCR and Genomic Sequencing in the Field. Tropical Medicine and Infectious Disease. 2022;7(1):6. doi:10.3390/tropicalmed7010006
- Huanyu Wang SJ. Next-Generation Sequencing for Infectious Diseases Diagnostics. Journel Article. Clinical Laboratory News. September 2021 2021;47(7):10-18.
- Bani Baker Q, Hammad M, Al-Rashdan W, Jararweh Y, Al-Smadi M, Al-Zinati M. Comprehensive comparison of cloud-based NGS data analysis and alignment tools. Informatics in Medicine Unlocked. 2020;18:100296. doi:10.1016/j.imu.2020.100296
Stephen Vella, PhD serves as a Medical Science Liaison for bioMérieux US Medical Affairs Division. His background is in Microbiology and has been working in diagnostics for approximately two years. His primary role serves to assist as neutral entity mediating scientific dialogue exchange for bioMérieux’s molecular and microbiology diagnostic portfolio.
About the Author

Stephen Vella Ph.D.
serves as a Medical Science Liaison for bioMérieux US Medical Affairs Division. His background is in Microbiology and has been working in diagnostics for approximately two years. His primary role serves to assist as neutral entity mediating scientific dialogue exchange for bioMérieux’s molecular and microbiology diagnostic portfolio