The role of cycle threshold values in infectious disease diagnostics
LEARNING OBJECTIVES
Upon completion of this article, the reader will be able to:
1. Recall the strengths and limitations of RT-PCR tests.
2. Describe the definition of cycle threshold (Ct) value and how it is used in infectious disease diagnostics.
3. Discuss research into the utility of Ct values in assessing COVID-19 disease progression.
4. Discuss research into the utility of Ct values in assessing disease progression for other respiratory diseases as well as gastrointestinal illnesses.
Effective interventions in the management of infectious diseases rely on rapid and specific detection of causative pathogens, driving patient care decisions and outcomes. Successful pathogen detection is facilitated by reliable, accurate diagnostic tools; and among the variety of testing tools available, molecular assays such as polymerase chain reaction (PCR) have become dominant. Sensitivity and specificity are paramount in diagnostics and molecular testing provides both. In theory, PCR can detect the presence of a single copy of nucleic acid from a single organism. The use of sequence-specific primers and probes in these assays yields unparalleled specificity and allows for testing to be multiplexed to detect multiple pathogens in one test run. This specificity allows not only for detection, but also for determining the presence of specific genes for typing different strains and antimicrobial resistance profiling.1 Alternatively, primers and probes can be designed to be less specific, or target highly conserved regions, to provide broad coverage and detect divergent pathogen genomes.2
PCR testing is successful in identifying organisms that cannot be grown in vitro or in situations where current culturing techniques lack sensitivity and/or require prolonged incubation periods.2 Other testing methods, such as culture or serology, often do not match the level of sensitivity or timeliness of molecular testing. In some cases, alternative methods may increase the risk of false negatives or incorrect results in critical testing situations where fastidious pathogens may be difficult to grow or immune responses difficult to detect.
Utility of cycle threshold values
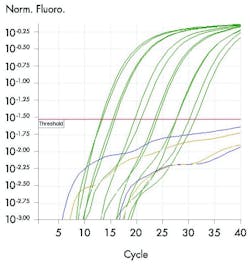
Effective PCR tests have now been developed for a wide variety of pathogens. However, these PCR tests, specifically real-time PCR (RT-PCR) tests, can reveal more than just the simple presence of a pathogen. Results from molecular tests are typically reported in a binary, positive or negative manner; however, this approach may not recognize the full benefits these assays have to offer. In addition to reporting pathogen detection, RT-PCR reports cycle threshold (Ct) values. The cycle threshold is defined as the thermal cycle number at which the fluorescent signal exceeds that of the background and, therefore, passes the threshold for positivity. Ct value is inversely related to the quantity of target nucleic acid in the sample, with lower Ct values reflecting higher viral loads, which can allow for comparison of relative levels of pathogen. Using this principle, a more specific quantification may be obtained with the use of a standard curve of known DNA quantity. This quantification can help establish pathogen burden and distinguish between what are normal, commensal levels and what are pathogenic levels of a microbe.
In addition to quantification, Ct values can provide additional information that may be useful when complex or unexpected results are received. Analysis of amplification curves and Ct values can be helpful in assessing sample quality and/or potential contamination. For patients with co-infections, the Ct values from a multiplex panel can help clarify which pathogen is most likely the causative agent in an illness. For patients on antimicrobial therapy, comparison of Ct values over time can be used as an indicator of the response to therapy.1
While PCR testing and Ct values can offer a deeper insight during the diagnostic process, they are not without limitations; perhaps the most important of which is the inherent inability to distinguish between live and dead organisms. The exquisite sensitivity of PCR allows for the detection of minute quantities of nucleic acid; however, this does not always correlate with the presence of live organisms. The assay will amplify nucleic acid from dead microbes as readily as from living.1 As dead organisms can be shed for weeks after recovery, a recovering patient might have the same positive test result as an actively infected patient. It is important to be able to distinguish between these two patients, as the positive test from the recovering patient may have no clinical relevance, while the actively infected patient may require treatment. In this situation, utilizing Ct values for relative quantification of the pathogen may help the clinician distinguish between these two patients.3
Ct values in COVID-19
The ongoing COVID-19 pandemic, with its clinical unknowns and a lack of standardized quantitative assays for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), has brought on an unprecedented level of interest in the clinical utility of Ct values. Patients infected with SARS-CoV-2 can display an array of clinical presentations, ranging from an absence of symptoms, to the requirement of intensive care and respiratory support, to an eventually fatal outcome.5
In fact, the proposed disease pathogenesis of COVID-19 indicates two overlapping disease states: an initial viremic phase where signs and symptoms are attributable to viral replication, and a subsequent inflammatory phase where the severity and critical illness is attributable to a “cytokine storm.” 6 While these two phases have different management implications from a treatment choice perspective – antiviral/antibody therapy versus anti-inflammatory therapy – a clear prognostic approach is still missing. The ability to predict prognosis and infectiousness of patients during the viraemic phase could fundamentally impact triage and management of patients and, potentially, of their contacts.
Early in the COVID-19 pandemic it was suggested that evidence from the 2002 SARS-CoV epidemic indicated that higher viral loads were associated with increased need for intensive care and overall worse outcomes. While there are differences between SARS-CoV and SARS-CoV-2, it has emerged that, similar to SARS-CoV, the viral load of SARS-CoV-2 could also play a key role in determining COVID-19 disease severity and prognosis, as well as infectiousness.3, 7-11
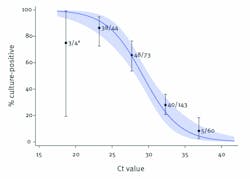
While the recent appearance of antigen tests has added to the panel of diagnostic solutions for COVID-19, RT-PCR has been the gold standard diagnostic approach since the beginning of the pandemic. Several of the COVID-19 RT-PCR solutions available on the market provide Ct values as part of their results data, and experts argue that these could be used as a semi-quantitative proxy for viral load in the absence of quantitative assays.Caption: The ability to recover infectious virus is strongly related to Ct value, as the estimated odds ratio of finding infectious SARS-CoV-2 virus decrease by 0.67 for every unit increase in Ct value. All five samples with Ct >35 from which virus was successfully propagated were from symptomatic patients and none had severe illness. Source: Centers for Disease Control and Prevention. Wide confidence levels is due to small sample size.
Source: CDC Library: COVID-19 Science Update: 09/04/2020
A recent systematic review assessed evidence to establish whether SARS-CoV-2 Ct values correlate with clinical outcomes and, therefore, whether they could provide valuable patient management information to clinicans.12 The findings indicate that lower Ct values may be associated with adverse outcomes and, in several instances, a correlation with clinical course and prognosis was identified. While the review concluded that, in general, higher Ct values correlate with lower viral loads, limitations have been highlighted around the fact that Ct values and lower viral load may not be directly proportional due to the presence of inhibitory factors within samples as well as the linear dynamic range of the assay.4
While the results of SARS-CoV-2 PCR diagnostic assays remain – in the majority of cases – reported as positive or negative results in the clinical setting, a discussion has emerged about the possibility of reporting and interpreting Ct values to aid clinicians in patient management. The increasing evidence supporting the utility of Ct values has led public health authorities, such as Public Health England and the Centers for Disease Control and Prevention (CDC), to develop guidance and recommendations regarding their use. Experts agree that if Ct values are to be reported, linearity, limits of detection and reference quantification curves must be standardized and validated. Interpretations of a single positive Ct value may be of limited use in the clinic.13, 14 However, indications are emerging that serial Ct values may offer greater utility for the purpose of clinical management.13
What is certain is that – in the absence of quantitative assays for SARS-CoV-2 – Ct values have become of increasing interest with regard to providing interpretative guidance, including as a potential benchmark for the development of antigen and rapid tests.Ct values beyond COVID-19
With, finally, a light at the end of the tunnel with the emergency use authorization (EUA) of several COVID-19 vaccines, it would be remiss not to highlight the potential application of Ct values beyond SARS-CoV-2. Prior to the COVID-19 pandemic, the most common causes of respiratory illness in seasonal circulation were influenza virus, respiratory syncytial virus (RSV), parainfluenza virus (PiV), human metapneumovirus (HMPV), adenovirus (AdV), rhinovirus and coronavirus (hCoV). RT-PCR is one of the most common diagnostic modalities for detection and identification of respiratory pathogens. There are several studies that have attempted to elucidate the clinical utility of Ct values for respiratory pathogens with contradictory reports that Ct values are associated with disease severity, intensive care unit (ICU) admission and length of hospital stay (LOS).15-17
The results of many studies evaluating associations between Ct values and clinical outcomes for patients with influenza were conflicting. However, a large influenza study identified that patients with low Ct values were significantly more likely to self-report moderate to high disease severity and fever.18 Several studies of patients with RSV reported association between low Ct values with clinically meaningful outcomes such as hospitalization, radiographical evidence of pneumonia or LOS, while others have failed to identify such associations. Despite many small rhinovirus studies having no association between Ct values and patient outcomes, there are a few large studies that found that low Ct values were associated with symptom severity and hospital LOS.19 For non-SARS-CoV-2 respiratory pathogens, Ct values for some respiratory infections may be useful in guiding treatment and healthcare decisions; however, the evidence supporting utility of Ct values is conflicting and warrants further investigation.
Multiplex PCR testing for viral, bacterial, and parasitic causes of gastrointestinal illnesses is one of the newer uses of syndromic diagnostic testing. As infectious diarrhea is estimated to cause more than 48 million illnesses and 3,000 deaths per year in the United States alone, any markers that could potentially help stratify patients to aid in clinical management is greatly needed. Compared to respiratory illness, there are fewer studies investigating a correlation of Ct values and clinical outcomes for gastrointestinal pathogens. A case-control study evaluated the application of molecular detection on relative amounts of gastrointestinal pathogens.20 There were significantly higher pathogen loads in cases versus controls for Campylobacter spp., Salmonella spp. Enterotoxigenic Escherichia coli (ETEC), typical Enteropathogenic E. Coli (EPEC) and C. parvus/hominis. Of note, Ct values for C. difficile and Enteroaggregative E.Coli (EAEC) were significantly lower for cases than for controls in patients 21–50 years old, and Ct values for atypical EPEC were significantly lower in cases than control for patients younger than 5 years old.
This study highlighted that low Ct values indicate high pathogen load and have the potential to suggest microbiological causes of gastroenteritis. A separate study specifically assessed the utility of Ct values on C. difficile disease and found significantly lower Ct values reported for patients with severe disease versus those with mild/moderate disease (median Ct values 25.9 vs 28.1, respectively; p=0.00001), with significant difference in the median age between samples with Ct values <25 and Ct values >25 , 79 and 74 years, respectively (p=0.004).21As treatment of C. difficile infections is guided by severity of symptoms and disease, the ability to predict severity of disease through Ct values as a surrogate for bacterial load has the potential to impact patient management and antimicrobial selection.
Conclusion
In conclusion, there are many considerations when utilizing Ct values as a surrogate for pathogen load in the laboratory setting and for use in the patient management decisions. However, the COVID-19 pandemic has highlighted the potential ability of Ct values to assess disease severity and those at higher risk of mortality. Although for other infectious pathogens the evidence is not as conclusive and many studies are conflicting, we should continue to evaluate the utility of Ct values to determine impact on patient management, as well as educate both laboratorians and clinicians on how to best interpret this numerical degree of PCR test positivity.
References
- Kralik P and Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol, 2017. 8:108. doi: 10.3389/fmicb.2017.00108.
- Yang S and Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis, 2004. 4(6): 337-48. doi: 10.1016/S1473-3099(04)01044-8.
- Tom MR and Mina MJ. To interpret the SARS-CoV-2 Test, consider the cycle threshold value. Clin Infect Dis, 2020. 71(16): 2252-2254. doi: 10.1093/cid/ciaa619.
- Aquino-Jarquin G. The raw Ct values from RT-PCR detection are not viral load quantitation units. Clin Infect Dis, 2020. doi: 10.1093/cid/ciaa830.
- Poletti P et al. Probability of symptoms and critical disease after SARS-CoV-2 infection. 2020. arXiv:2006.08471.
- Gandhi RT, Lynch JB and Del Rio C. Mild or moderate Covid-19. N Engl J Med, 2020. 383(18): 1757-1766. doi: 10.1056/NEJMcp2009249.
- Cheng VC, et al. Viral replication in the nasopharynx is associated with diarrhea in patients with severe acute respiratory syndrome. Clin Infect Dis, 2004. 38(4): 467-75. doi: 10.1086/382681.
- Chu CM, et al. Initial viral load and the outcomes of SARS. CMAJ, 2004. 171(11): p. 1349-52. doi: 10.1503/cmaj.1040398.
- Geddes L. Puzzle over viral load. New Sci, 2020. 245(3276): 8. doi: 10.1016/S0262-4079(20)30658-8.
- Joynt GM and Wu WK. Understanding COVID-19: what does viral RNA load really mean? Lancet Infect Dis, 2020. 20(6): p. 635-636. doi: 10.1016/S1473-3099(20)30237-1.
- Ng EK, et al. Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin Chem, 2003. 49(12): p. 1976-80. doi: 10.1373/clinchem.2003.024125.
- Rao SN, et al. A systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther. 9(3): 573-586. doi: 10.1007/s40121-020-00324-3.
- Understanding cycle threshold (Ct) in SARS-CoV-2 RT-PCR - Understanding_Cycle_Threshold__Ct__in_SARS-CoV-2_RT-PCR_.pdf. 2020. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/926410/Understanding_Cycle_Threshold__Ct__in_SARS-CoV-2_RT-PCR_.pdf.
- Frequently asked questions about Coronavirus (COVID-19) for laboratories. Centers for Disaease Control and Prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/faq.html. Accessed January 11, 2021.
- Feikin DR, et al. Association of higher MERS-CoV Virus load with severe disease and death, Saudi Arabia, 2014. Emerg Infect Dis, 2015. 21(11): 2029-35. doi: 10.3201/eid2111.150764.
- Hasegawa K, et al. Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: multicenter cohort studies in the United States and Finland. J Infect Dis, 2015. 211(10): 1550-9. doi: 10.1093/infdis/jiu658.
- Wishaupt JO, et al. Pitfalls in interpretation of CT-values of RT-PCR in children with acute respiratory tract infections. J Clin Virol, 2017. 90: 1-6. doi: 10.1016/j.jcv.2017.02.010.
- Spencer S, et al. Factors associated with real-time RT-PCR cycle threshold values among medically attended influenza episodes. J Med Virol, 2016. 88(4): 719-23. doi: 10.1002/jmv.24373.
- Clark TW, et al. Viral load is strongly associated with length of stay in adults hospitalised with viral acute respiratory illness. J Infect, 2016. 73(6): 598-606. doi: 10.1016/j.jinf.2016.09.001.
- Bruijnesteijn van Coppenraet LE, et al. Case-control comparison of bacterial and protozoan microorganisms associated with gastroenteritis: application of molecular detection. Clin Microbiol Infect, 2015. 21(6): 592 e9-19. doi: 10.1016/j.cmi.2015.02.007.
- De Francesco MA, et al. Correlation between tcdB gene PCR cycle threshold and severe Clostridium difficile disease. Anaerobe, 2019. 59: 141-144. doi: 10.1016/j.anaerobe.2019.06.0
About the Author

Sonia Rao, PharmD, BCIDP, BCPS
is a Senior Medical Science Liaison at QIAGEN.

Davide Manissero, MD, MRCPCH, MSc, DTM&H
is a Vice President and Head of Medical Affairs and Chief Medical Officer – Infection and Immune Diagnostics – at QIAGEN.

Kelly Hughes, PhD
is a Medical Science Liaison in infectious diseases for Medical Affairs at QIAGEN.