The hard truth about loose stool— C. difficile diagnostics in a new era
Discovery of disease and matching specific microorganisms to the disease they cause has been key for medicine. The roots of this process date back to 460 BC, to Hippocrates, a Greek physician, who postulated that “bad air” was to blame for disease. As improvements to microscopes, technology, and description of anatomy advance, so too does the understanding of disease.
C. difficile—a brief history in time
For over a century now, the aspects surrounding the clinical description, and subsequent diagnosis, of Clostridium difficile infection (CDI) have existed. John Finney, a surgeon at Johns Hopkins Hospital, is among the first to document the clinical context of CDI in a patient who had undergone a gastroenterostomy for an ulcerated pylorus in 1893.1 The patient ultimately expired 15 days after surgery where an autopsy performed on the patient revealed “pseudodiphtheritic enteritis” involving the ileum and colon.
In 1935, Hall and O’Toole examined the early invasion of the intestinal tract of infants describing Bacillus difficilis as part of normal healthy flora in neonates.2 The association between antibiotic use and C. difficile antibiotic-associated diarrhea (CDAD) was further described by Reiner and colleges in a case of pseudomembranous colitis following use of aureomycin and chloramphenicol.3 This finding fostered a perspective of CDI, incorrectly attributable to “staphylococcal enterocolitis,” which was a prevailing dogma of the 1950s.4,5 Subsequent evaluation demonstrated that “staphylococcus colitis” cases were not associated with pseudomembranes, nor were animal genuine cases of pseudomembranous colitis associated with staphylococci. The failure to establish “staphylococcus colitis” in animal models shifted the perspective of the disease away from Staphylococcus aureus and toward C. difficile toxin production, which was demonstrated by Bartlett and colleagues in 1977 using antibiotic-induced colitis hamster models.6 The role of toxin-mediated disease, and the basis of free toxin testing, was later established by Tracy Wilkins, who successfully purified the toxins in 1982.7,8 However, it required nearly 30 years to definitively establish the role of both toxin A and toxin B in CDI8, which fames our current understanding of CDI as a toxin-mediated disease. Today, we continue to scope contemporary challenges, which include the role of asymptomatic carriage, defining variables involved in transmission, and response to new therapies, even leading to the reclassification of C. difficile from Clostridium difficile to the taxonomically valid Clostridiodes difficile.
Diagnostic dilemmas and pathways for testing
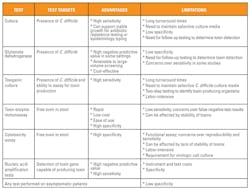
Despite increased understanding of the pathogenesis of CDI, many questions regarding the optimal diagnostic approach to CDI exists and several laboratory tests, targeting different diagnostic targets and approaches, are used. Toxigenic culture remains the gold standard assay for detection of toxigenic C. difficile.
Cytotoxin assays have historically been acknowledged as the “gold standard” approach to the diagnosis of CDI. Cytotoxin neutralization utilizes tissue-culture techniques as a laboratory test, which directly assays for a cytotoxic phenotype in prepared stool specimens. Neutralization of toxin (cytotoxic effect) facilitates a highly specific test in the setting of symptomatic patients. However, over time, lack of sensitivity of cytotoxin neutralization assays and the length of time needed to provide a laboratory result have led experts to question the role of cytotoxin neutralization as a representative gold standard reference.9 Direct detection of toxin in clinical samples is often subject to thermostability issues because toxins are biologically stable across temperature ranges, including body temperatures, which may naturally affect stability of toxin testing prior to testing.10 Although highly specific, cytotoxin assays lack sensitivity compared to toxigenic culture, which is tested by culture of C. difficile from stool on selective media and then assaying for toxigenic potential of colonies recovered in culture. Compared to toxigenic culture, cytotoxin neutralization assays achieve only 60–85 percent sensitivity.11,12
For a number of clinically important reasons, enzyme immunoassays (EIAs) replaced cytotoxin neutralization and toxigenic cultures as the preferred diagnostics for CDI. EIAs targeted against toxins A/B provided a rapid laboratory response to suspected CDI, which could be easily performed by laboratory staff and provided highly specific and cost-effective laboratory testing. Support for these clinical goals often counterbalanced requirements for sensitive testing as many EIAs were validated compared to cytotoxin neutralization assays, which we now acknowledge as an insensitive gold standard. While some studies with EIAs compare favorably (>95 percent sensitivity) to fibroblast cytotoxicity neutralization assays,13 others have shown wide ranging sensitivities of between 53–90 percent.14 However, one must account for the difference in sensitivity between cytotoxin assays and toxigenic culture when interpreting these data sets. Cytotoxicity assays lack sensitivity compared to toxigenic culture, thus studies describing 90 percent positive predictive agreement between EIA testing and cytotoxicity assays actually represent much lower sensitivity when compared to toxigenic culture, which have been reported at only 65 percent sensitivity compared to toxigenic culture.13
Questions surrounding the sensitivity of EIA testing led many laboratories and hospitals using EIA testing to provide EIA testing in triplicate with each order for C. difficile to increase sensitivity, and reports of poorly performing EIAs in symptomatic patients were reported.15
Glutamate dehydrogenase (GDH) is another form of testing that has been successfully incorporated into contemporary testing algorithms. GDH is an enzyme produced by various bacteria essential to the metabolic conversation of glutamate to -ketoglutarate. GDH screening has proved to be a sensitive, cost-effective screen for the presence of C. difficile direct from stool specimens.16 GDH screening requires additional testing in the setting of positive results because GDH cannot detect the presence of toxin or toxigenic organisms. A positive GDH result reflexes to testing with EIA or molecular assays that can determine the toxin potential of the specimen. Thus, the performance and clinical confidence in GDH algorithms depend on the form of follow-up testing used. Some laboratories have adopted three-tier approaches to GDH screening, which utilize GDH testing, followed by EIA testing, and finally PCR testing for EIA-negative cases. As most specimens submitted for testing are actually negative, GDH screening offers the advantages of high-volume screening which is cost-effective compared to testing all specimens by PCR.17 However, multistep algorithms add complexity to patient reporting. Multiple assays are used to provide different results and reporting may be delayed compared to single-testing strategies, which in some settings have made two- or three-step testing algorithms impractical. Performance of GDH as a screening assay has been reported between varied between 97-83 percent in some studies,17,18 raising the question, is GDH sufficiently sensitive to provide a high predictive value needed for rule-out testing?
Concerns over insensitive testing for CDI led to the molecular movement away from EIA (or GDH) testing for the molecular detection of the toxin B gene. Detection of the gene correlates with an organism capable of producing toxin. Molecular assays report much higher sensitivities than EIA testing.19 Molecular-based testing was anointed the test of choice among many experts and healthcare institutions. However, as molecular testing gained prominence over the last decade, it has also generated new clinical and laboratory questions.
How does asymptomatic colonization affect one’s choice of testing options?
Patients who asymptomatically carry toxigenic C. difficile in the intestinal flora have been recognized for well over 50 years.20 Asymptomatic C. difficile colonization is common and present in up to 30 percent of hospitalized patients. Thus, submitting laboratory testing on patients whose diarrhea is not likely attributable to C. difficile is problematic, and non-specific, regardless of the laboratory assay used. Molecular assays with higher sensitivity are more likely to report positive results in a higher proportion of these patients than EIA testing.
Is “free-toxin” testing a better approach to the diagnosis of CDI?
There is little question that cases of CDI can test false negative by EIA and false-negative toxin results are risk factors for poor outcomes and death in patients with fulminant CDI.21 Even those who advocate for EIA approaches report deaths of patients who test negative by EIA testing compared to PCR.22 These observations have led to conclusions that toxin assays cannot rule out the possibility of CDI.23 The frequency with which these cases are reported remain an area of debate. Physicians and laboratorians who have failed to diagnose CDI in patients are shaped by their experiences. Others feel that anecdotal experiences are an overestimation of the scope of EIA false-negative testing. Three independent studies, conducted in the UK, Canada, and the U.S., concluded that toxin assays correlate stronger to the diagnosis of CDI than PCR results do and that reliance on PCR may overcall many diagnoses of CDI that are attributable to other causes of diarrhea.24,25 As a result, some experts consider EIA-negative/PCR-positive results likely represent colonization.9
How do we reconcile these two competing discussions?
The difficulty in establishing the specificity (and reducing false positives) for CDI is founded in relativity that symptoms associated with CDI vary widely from a spectrum that includes frequent watery diarrhea to fulminant life-threatening pseudomembranous colitis. It is this difficulty in recognizing symptoms attributable to definitively toxigenic C. difficile compared to other infectious, and non-infectious, causes of diarrhea among hospitalized patients that create challenges for clinicians. Clinical order sets reminding ordering physicians of the frequency and intensity of diarrhea, other possible causes of diarrhea such as laxatives or feeding tubes, and objective clinical markers of CDI, such as leukocytosis and abdominal cramping, all help to focus testing and increase the pre-test probability of disease within laboratory orders. Clinical recognition that we treat the patient and not the test result is an important message.26 This message underscores that the diagnosis of CDI is not made by a laboratory assay, but rather that laboratory testing should provide high-fidelity results used to support clinical perspective. Simply put, laboratories need to communicate the strengths and pitfalls of various testing modalities to all groups involved and assess the relative risks of missing a diagnosis of CDI versus the inability to control and steward C. difficile orders leading to false-positive reporting.
Can new testing options provide clarity to a complicated diagnostic scene?
Limitations and challenges surround aspects of current C. difficile assays making it difficult for clinicians and laboratory personnel to fully understand the benefits or limitations of various diagnostic options. GDH-based testing requires multistep procedures and reporting that can complicate and often delay reporting compared to standalone testing. PCR-based assays can create false positives when test ordering cannot be controlled or limited to symptomatic patients, and often represent expensive testing options. EIA testing, assays for free toxin, although quick and cost-effective, create concerns over poor sensitivity and are subject to biological variability as a result of liable toxin targets.
Single Molecule Counting, linked with traditional immunoassay technology, has made it possible to bridge the challenges posed by today’s diagnostic options for CDI. Findings from the first fully-automated, in-vitro diagnostic to employ Single Molecule Counting technology—which provides capture of microparticles coated with antibodies targeting C. difficile toxins A and B at levels down to picogram-per-milliliter levels—are promising. The surface chemistry of the microparticles increases the affinity of the antibody capture, and the microparticles are magnetically separated and washed to remove unbound proteins and nonspecific binding. Fluorescent dye-labeled antibodies trap captured toxin converting the process into a detectable signal. Each signal corresponds to a single analyte molecule. The signals are illuminated by a laser, resulting in a distinct and accurate flash of light. Single Molecule Counting offers the possibilities of highly sensitive, free-toxin testing at sensitivity levels that rival PCR. Recent data presenting at both the 2018 European Congress of Clinical Microbiology and Infectious Diseases and the American Society of Microbiology Microbe annual meetings indicates that the toxin A/B assays using Single Molecule Counting technology yielded 98 percent sensitivity and 100 percent specificity compared to multistep PCR-and toxin testing. Compared to cell cytotoxicity neutralization testing, the new assays detected 97.7 percent of toxin-positive stool samples and detected toxins in 23 percent of PCR-positive and CCNA-negative samples.27 These preliminary findings indicate that the assay may offer a standalone solution for CDI diagnostics.
While the stability of single-molecule testing for C. difficile-associated toxins requires further investigation, these preliminary findings suggest that the detection of free toxin in a highly automated platform capable of successfully ruling in or out toxin-producing C. difficile in clinical samples is an attainable goal. Combining highly sensitive testing with specific toxin detection allows physicians to target testing to symptomatic patients, knowing laboratory testing can successfully catch the diagnosis of CDI while providing high negative predictive value that can confidently rule out CDI, a variable that has been questioned with traditional EIA testing.
While debates over laboratory testing for CDI are likely to continue, the availability of an assay that can provide highly sensitive, specific, rapid, and affordable diagnosis for CDI is a goal we can all agree on, further validating the perspective of Robert Koch’s offered over a century ago. “As soon as the right method was found, discoveries came as easily as ripe apples from a tree.”
REFERENCES
- Finney J MT. Gastroenterostomy for cicatrizing ulcer of the pylorus. Bull Johns Hopkins Hosptial. 1893;4:53–5.
- E H; IO. Intestinal flora in new-born infants with a description of a new pathogenic anaerobe, bacillus difficilis. Am Jounal Dis Child. 1935;49(2):390–402.
- Reiner L, Schlesinger KJ, Miller G. Pseudomembranous colitis following aureomycin and chloramphenicol. AMA Arch Pathol. 1952 Jul;54(1):39–67.
- Bartlett JG, Gorbach SL. Pseudomembranous enterocolitis (antibiotic-related colitis). Adv Intern Med. 1977;22:455–76.
- ANTIBIOTICS, staphylococcal enteritis and pseudomembranous enterocolitis. N Engl J Med. 1953 Jul;249(1):37–40.
- Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis. 1977;136(5):701–5.
- Sullivan NM, Pellett S, Wilkins TD. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun. 1982;35(3):1032–40.
- Lyerly DM, Lockwood DE, Richardson SH, Wilkins TD. Biological activities of toxins A and B of Clostridium difficile. Infect Immun. 1982;35(3):1147–50.
- Wilcox MH, Planche T, Fang FC, Gilligan P. What is the current role of algorithmic approaches for diagnosis of Clostridium difficile infection? J Clin Microbiol. 2010;48(12):4347–53.
- Chang TW, Lauermann M, Bartlett JG. Cytotoxicity assay in antibiotic-associated colitis. J Infect Dis. 1979;140(5):765–70.
- Binning M, John MA, Schieven BC, Austin TW, Lannigan R, Hussain Z. Comparison of culture, cytotoxin assay and two EIA tests with clinical diagnosis of Clostridium difficile-associated diarrhea. Can J Infect Dis. 1994;5(4):163–7.
- Reigadas E, Alcalá L, Marín M, Muñoz-Pacheco P, Catalán P, Martin A, et al. Clinical significance of direct cytotoxicity and toxigenic culture in Clostridium difficile infection. Anaerobe. 2016;37:38–42.
- Merz CS, Kramer C, Forman M, Gluck L, Mills K, Senft K, et al. Comparison of four commercially available rapid enzyme immunoassays with cytotoxin assay for detection of Clostridium difficile toxin(s) from stool specimens. J Clin Microbiol. 1994;32(5):1142–7.
- Turgeon DK, Novicki TJ, Quick J, Carlson L, Miller P, Ulness B, et al. Six rapid tests for direct detection of Clostridium difficile and its toxins in fecal samples compared with the fibroblast cytotoxicity assay. J Clin Microbiol. 2003;41(2):667–70.
- Manabe YC, Vinetz JM, Moore RD, Merz C, Charache P, Bartlett JG. Clostridium difficile colitis: an efficient clinical approach to diagnosis. Ann Intern Med. 1995;123(11):835–40.
- Quinn CD, Sefers SE, Babiker W, He Y, Alcabasa R, Stratton CW, et al. C. Diff Quik Chek complete enzyme immunoassay provides a reliable first-line method for detection of Clostridium difficile in stool specimens. J Clin Microbiol. 2010;48(2):603–5.
- Novak-Weekley SM, Marlowe EM, Miller JM, Cumpio J, Nomura JH, Vance PH, et al. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010;48(3):889–93.
- Larson AM, Fung AM, Fang FC. Evaluation of tcdB Real-Time PCR in a Three-Step Diagnostic Algorithm for Detection of Toxigenic Clostridium difficile. J Clin Microbiol. 2010;48(1):124–30.
- Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, et al. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010;48(10):3719–24.
- Bartlett JG, Onderdonk AB, Cisneros RL. Clindamycin-associated colitis in hamsters: protection with vancomycin. Gastroenterology. 1977;73(4 Pt 1):772–6.
- Dallal RM, Harbrecht BG, Boujoukas AJ, Sirio CA, Farkas LM, Lee KK, et al. Fulminant Clostridium difficile: an underappreciated and increasing cause of death and complications. Ann Surg. 2002;235(3):363–72.
- Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med. 2015;175(11):1792–801.
- Fang FC, Polage CR, Wilcox MH. Point-Counterpoint: What Is the Optimal Approach for Detection of Clostridium difficile Infection? J Clin Microbiol. 2017;55(3):670–80.
- Longtin Y, Trottier S, Brochu G, Paquet-Bolduc B, Garenc C, Loungnarath V, et al. Impact of the type of diagnostic assay on Clostridium difficile infection and complication rates in a mandatory reporting program. Clin Infect Dis. 2013;56(1):67–73.
- Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13(11):936–45.
- Dubberke ER, Burnham C-AD. Diagnosis of Clostridium difficile Infection: Treat the Patient, Not the Test. JAMA Intern Med. 2015;175(11):1801–2.
- Sandlund J, Bartolome A, Bishop J. Clinical Evaluation of the Singulex Clarity C. diff Toxins A/B Assay, Currently in Development, for Ultrasensitive Detection of Clostridium difficile Toxins A and B. In: European Congress of Clinical Microbiology and Infectious Diseases Annual Meeting. 2018.