Microbiology, molecular, MALDI-TOF or all of the above?
While hematology and clinical chemistry labs were automated many years ago, the microbiology lab is still largely a hands-on process. In part, this reflects the complexity of microbiology. There are hundreds of infectious pathogens, including bacteria, viruses, fungi, and parasites. In order to properly diagnose these pathogens, they have to be cultured, so the lab has enough of the pathogen to identify it via a litany of biochemical tests. Because all of these bugs are so different, some take just hours to grow, while certain fungal infections can take weeks. Some bacteria are aerobic, while others are anaerobic. All have ideal growth media to foster optimal growth. These “pre-analytical” considerations are vital to rapid identification. In other words, the microbiology lab is as complex as microbiology. No two cases are exactly the same, so automating the microbiology lab is a much more
difficult challenge.
Molecular and MALDI-TOF
The newest technologies in the microbiology lab hold the potential to solve many of these problems. Molecular techniques can generate results and positive identifications remarkably quickly; so can Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) mass spectrometry, which is having a significant impact on clinical microbiology.
Automated blood culturing, antimicrobial susceptibility testing (AST), and MALDI-TOF systems lead to more accurate and faster results, with less wasted materials and tech time. Mass spectrometry has a simple sample preparation process and generally requires a smaller amount of culture growth for identification. MALDI-TOF generates high throughput of dozens of pathogens per run. Automated ID/AST provides a broad range of Gram-negative AST cards, and, in some cases, three-drug yeast susceptibility testing on a single card with results in under eight hours. Multiplexing PCR systems have evolved from identifying a single organism to identifying groups of bacteria, viruses, and yeast that often lead to certain conditions such as sepsis, all in one run lasting less than 60 minutes. Some systems can also identify sequences of nucleotides that are unique to dominant types of antibiotic resistance, such as methicillin, vancomycin, and carbapenem resistance. Such features can speed up treatment decisions and may be able to facilitate targeted therapy sooner.
However, molecular and MALDI-TOF are still held hostage to the pre-analytical steps and delays. PCR techniques—even multiplexing techniques that search for likely pathogenic suspects based on the “syndromic” symptoms of the patient—still must wait for culturing. Likewise, MALDI-TOF requires cultured organisms, although less growth is often acceptable for identification by mass spectrometry than is required for standard biochemical
techniques.
A holistic approach
A new generation of pre-analytical systems that require very little interaction and improve error rates and inefficiencies holds the promise of maximizing the benefits of the new analytical systems and creating a new holistic system—from sample collection to results reporting— that is more than the sum of its many parts.
Sometimes called “walk-away” specimen pre-analytical processing systems, these new tools include a new generation of automated plate streakers, Gram slide preparation, enrichment broth inoculation, and newer tools including the digital imaging and magnification of growth colonies that reduce the time required to identify bacterial growth. This feature, in particular, could prove to be very synergistic with MALDI-TOF systems, which generally require less growth for positive identification. The microbiology lab is on the brink of complete automation that encompasses all aspects of specimen processing and management, incubation, digital colony selection, and software to assist laboratorians in analysis and results reporting. Some full lab automation systems include algorithms and artificial intelligence that automatically read, interpret, and segregate bacterial cultures.
Rapid and robust pre-analytics are vital to maximizing the benefits of the new identification systems that have changed the face of the microbiology lab over the last decade. These benefits can be lost if the culture process is slow, inefficient, or prone to errors. Think of it this way: a best-in-class automobile designed for luxury, speed, and fuel efficiency is useless on a road riddled with sink holes, water damage, and debris. So it’s vital to pair cutting-edge diagnostic technology with cutting-edge pre-analytical processes and technologies.
New tools are also helping to bring new efficiencies to the identification process by assisting in the steps required for identification. For example, a laser trace tool can help lab staff select the precise growth cluster identified on a digital image. This cluster may be so small that finding and selecting it manually is a major challenge. However, by using a laser trace, pinpointing the optimal clusters should be much easier, which, in turn, allows for optimal MALDI-TOF identification. Artificial intelligence and reading algorithms can integrate with chromogenic culture media to automatically detect the color change that indicates positive growth. Plates that are negative can be automatically discarded by the system, while those that are positive are expedited for susceptibility testing.
Creating synergies
The ideal marriage between pre-analytic automation and new identification techniques should enable the automatic processing of a sample during many of the steps inherent to microbiological analysis.
For example, the manual streaking and incubation processes should be automated and standardized by automated plate streaking systems and an innovative incubation which includes a growing list of automated features. Digital imaging and analysis can reduce the time needed to identify bacterial growth. These instruments can integrate with a lab’s automated blood culturing and antibiotic susceptibility testing. And algorithms and artificial intelligence automatically read, interpret, and segregate plates. Such systems hold the potential to move current laboratories to the digital microbiology era through high-resolution culture plate imaging, improving speed, interpretation, reliability, and accessibility of results.
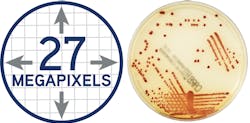
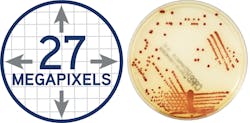
The ideal image acquisition technology should provide highly sophisticated lighting and a camera system to make each plate image clear and accurate—just like using a plate microscope with every plate, so lab staff can reliably make accurate work-up decisions. The lab camera optics should provide clear colony views, no matter the agar thickness. In Figure 1 (page 28), for example, the resulting usable image of this plate is 27 megapixels, which allows for the visualization of colonies even smaller than 0.1. This larger-than-life image can help techs detect growth invisible to the naked eye on the actual plate. After a set time, file images are compressed for storage.
Lab imaging systems should capture high resolution images of plates before incubation to automatically identify and eliminate any existing artifact. This high-resolution image approach for differential image analysis is a reliable warranty for accurate pathogen growth detection. Other key features should include high-throughput reading and the ability to quickly discard negative plates. This allows staff to focus on critical positive samples quickly and frees highly experienced laboratorians to do high-value work.
No two microbiology labs are the same. Ultimately, each must make choices that significantly enhance its operational efficiency, compensate for the growing shortage of specialized staff, and obtain the accreditation it needs to operate. This can be accomplished by streamlining workflows while delivering faster and more standardized results and improving traceability of analyses. The key for all labs is to find open, fully automated, modular and flexible diagnostic solutions that fit the unique needs of the lab and its patient population.
Richard Cleveland serves as Sr. Marketing Manager, U.S. Clinical Laboratory Optimization, for bioMerieux, Inc., provider of the VIRTUO and VITEK automated microbial identification systems.
Amanda Schmidt serves as Marketing Manager, COPAN Diagnostics, provider of the WASP DT and WASPLab microbiology specimen processors. In 2015, bioMerieux and COPAN began a partnership for integrating pre-analytic automation with various culturing and identification technologies. In January 2017, the COPAN-bioMerieux offering became available in the U.S.