To take test online go HERE. For more information, visit the Continuing Education tab.
LEARNING OBJECTIVES
Upon completion of this article, the reader will be able to:
1. Discuss the current outbreak of drug-resistant Pseudomonas aeruginosa.
2. Describe the background and epidemiology of carbapenem-resistant P. aeruginosa (CP-CRPA).
3. Differentiate between different testing methods for CP-CRPA.
4. List the entities and guidelines involved to combat future carbapenem-resistant P. aeruginosa (CP-CRPA) outbreaks.
In February 2023, both the Centers for Disease Control and Prevention (CDC) and the U.S. Food and Drug Administration (FDA) issued warnings to consumers and healthcare providers about infections due to an extensively drug-resistant Pseudomonas aeruginosa linked to the use of artificial tears.1,2 Sixty-eight patients in 16 states have been identified, with various clinical presentations including keratitis, respiratory and urinary tract infections, and sepsis. Patient outcomes included permanent vision loss, hospitalization, and one death with a bloodstream infection.
The P. aeruginosa isolates were initially identified by clinical laboratories and submitted to local public health laboratories (PHLs) and/or the CDC for additional antimicrobial susceptibility testing (AST) by reference broth microdilution and molecular characterization. Isolates associated with the outbreak were resistant or showed intermediate susceptibility to nearly all agents routinely tested and used clinically for pseudomonal infections, including cefepime, ceftazidime, aztreonam, imipenem, meropenem, piperacillin-tazobactam, ceftazidime-avibactam, ceftolozane-tazobactam, ciprofloxacin, amikacin, gentamicin, and tobramycin. The only agents that showed any activity in vitro were cefiderocol and colistin.
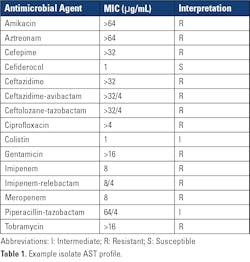
Even more alarming, was the finding that outbreak isolates carried two transmissible genes for the carbapenemases, VIM and GES, a combination not previously identified in carbapenemase-producing carbapenem-resistant P. aeruginosa (CP-CRPA) in the United States. Therapeutically, carbapenem resistance in P. aeruginosa due to carbapenemases are challenging, as CP-CRPA are likely to exhibit resistance to other b-lactam agents and newer b-lactam–b-lactamase combination agents, including those designed to treat complicated pseudomonal infections, such as ceftolozane-tazobactam.3,4 An example AST profile of one of the outbreak isolates is shown in Table 1.
This outbreak reaffirmed the importance of the clinical laboratory in the accurate detection of CRPA to help guide patient treatment, assist infection preventionists to help stop the local spread of CRPA, and support public health initiatives to combat antimicrobial resistance.
Background and epidemiology of CRPA
P. aeruginosa is commonly found in environmental sources, like soil and water. It is frequently associated with hospital-acquired infections and is especially problematic in immunocompromised individuals.5 Antimicrobial resistance in P. aeruginosa, as with other gram-negative bacteria, continues to increase globally and has been elevated as an agent of Serious Threat by the CDC and a Critical Priority pathogen by the World Health Organization (WHO).6,7 Of particular importance is carbapenem resistance, which can be due to the production of one or more carbapenemase enzymes (e.g., VIM, KPC, IMP, NDM). In contrast to carbapenemase production in Enterobacterales such as Klebsiella pneumoniae where KPC is the most prevalent carbapenemase in the United States, VIM is the most common carbapenemase in CP-CRPA.8,9 Carbapenem resistance in P. aeruginosa can also be due to non-carbapenemase mechanisms such as changes in membrane permeability or activation of efflux pumps and/or increased cephalosporinase activity. Some CRPA may result from a combination of carbapenemase and non-carbapenemase resistance mechanisms. While containing the spread of all CRPA is important, the quick detection and control of CP-CRPA is paramount because genes coding for carbapenemases, which typically reside on plasmids with other antimicrobial resistance genes, are easily spread among bacteria in healthcare settings and other environments.6
Laboratory identification of CRPA and CP-CRPA
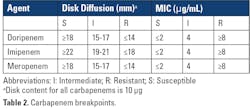
Results from routine AST performed in the clinical laboratory are used to identify CRPA. As the name implies, CRPA are resistant to one or more carbapenems (doripenem, imipenem, and/or meropenem) when using current breakpoints as listed in Clinical and Laboratory Standards Institute (CLSI) M100 33rd Edition Standard (Table 2).10Further characterization to determine if carbapenem resistance is due to carbapenemase production may be warranted for institutional infection control measures and/or epidemiological purposes. In some cases, clinicians may request the laboratory test for carbapenemase production in a CRPA isolate, as insight into the resistance mechanism may further guide patient management, although fewer than 5% of CRPA reported by the CDC have been shown to produce a carbapenemase.11 However, a recent study showed that an intermediate or resistant result for cefepime, ceftazidime and/or ceftolozane-tazobactam can be used as a signal that a CRPA isolate may produce carbapenemase.11 When testing 5,394 CRPA (resistant to imipenem or meropenem), only 177 (3%) were confirmed carbapenemase producers. Of the 903 isolates that were also intermediate or resistant to ceftolozane-tazobactam, 223 (25%) produced carbapenemase. Intermediate or resistant results for cefepime or ceftazidime can also be used to identify CRPA that are suspicious for carbapenemase production (sensitivity of 91%), but may be less useful given their much lower specificity for detecting CP-CRPA (50%) compared to ceftolozane-tazobactam (86%). One conclusion from these studies was that contemporary CRPA isolates in the United States that are susceptible to all three agents are unlikely to be carbapenemase producers. Some laboratories may use this criterion to determine which CRPA to further test for carbapenemase.
Phenotypic methods and genotypic methods can be used for detecting carbapenemases in CRPA. Phenotypic methods, such as the Modified Carbapenem Inactivation Method (mCIM), will tell whether or not a carbapenemase is being produced but cannot identify the specific carbapenemase or carbapenemase gene(s).12 Genotypic tests, on the other hand, mostly involve polymerase chain reaction (PCR) methods that target specific carbapenemase gene(s), and it should be assumed that an isolate that harbors a carbapenemase gene produces or is capable of producing the respective carbapenemase enzyme. The commercial genotypic methods used most commonly in clinical laboratories target the main carbapenemase genes reported to date in the United States, sometimes referred to as the “big 5”: KPC, NDM, VIM, IMP, and OXA-48.13 Consequently, the GES carbapenemase concomitantly present with VIM in the CP-CRPA responsible for the outbreak described above would not have been detected with most commercial molecular kits. In addition to PCR methods, at least one commercial lateral flow immunoassay detects the five most common carbapenemases and is gaining wide use in clinical laboratories.14 In contrast to genotypic methods, phenotypic methods may identify novel carbapenemases that are not targeted in genotypic assays. When a phenotypic test is positive but a genotypic test for the “big 5” is negative, methods such as whole genome sequencing may be used to further characterize the carbapenemase.
As some genotypic tests for carbapenemases can be performed directly from a clinical specimen (e.g., rectal swab or positive blood culture), the results that a carbapenemase gene is present may be available before carbapenem resistance is detected by a standard AST method. In such cases, it should be presumed that a positive result by a molecular method infers carbapenem resistance. CLSI M100 provides guidance for testing and reporting results in laboratories that use molecular methods, with or without a phenotypic test, for detecting carbapenem resistance. Specifically, Appendix H - Using Molecular Assays for Resistance Detection, contains this information, including how to handle discrepancies between phenotypic and genotypic results.10
Surveillance for CRPA and the Antibiotic Resistance Laboratory Network.
In 2016, the CDC established the Antibiotic Resistance Laboratory Network (AR Lab Network) to better understand the scope of antimicrobial resistance in the United States.8 The network consists of PHLs in all 50 states, several large cities, and Puerto Rico, and includes seven regional laboratories. The goal of the AR Lab Network is to support laboratory testing to detect and contain antimicrobial resistance in health care, the community, and the environment.
A major focus of the AR Lab Network has been carbapenem-resistant organisms including CRPA. PHLs have developed various mechanisms for interacting with their local clinical laboratories to obtain data and/or isolates. In some states with low prevalence of carbapenem resistant organisms (CRO), clinical laboratories may be required to submit all CRO to their public health laboratory. In others, where the burden of CRO is high, clinical laboratories may be required to report all CRO and submit only select species of CP-CRO, such as CP-CRPA or CP-CRO that produce less common carbapenemases. The AR Lab Network and local/state PHLs can assist clinical laboratories with identification and characterization of CRPA and/or CP-CRPA as well as other CROs because they often have additional testing capabilities beyond what is available in routine clinical laboratories, such as ability to strain type and perform whole genome sequencing. At the time of this writing, CDC’s AR Lab Network dashboard reports that during 2018–2021, there were 56,016 CRPA isolates tested, and 2.11% (N=1,181) had at least one targeted carbapenemase gene detected. The most common carbapenemase detected was VIM, found in 57.8% of CP-CRPA.11 This data was corroborated in a recent global, observational study of CRPA, which demonstrated differences in both prevalence and diversity of carbapenemases across various geographical regions. In the United States, only 2% (N=10) of CRPA isolates harbored a carbapenemase. Additionally, there was an increase in mortality in patients with CP-CRPA infections compared to those with non-CP-CRPA. This study highlighted the impact these pathogens have on clinical outcomes and importance of continued surveillance to monitor global patterns of carbapenemases and regional shifts in carbapenemase emergence.3
Laboratories should refer to local or state PHL guidelines for reporting of suspected or confirmed CRPA and potential isolate submission requirements. This is of particular importance when there is a suspected outbreak being investigated, as was the case with the VIM-GES-CRPA cases.
What is next for CRPA?
The continuing evolution of carbapenem resistance in P. aeruginosa worldwide highlights the ongoing need for various strategies and diligent efforts to combat CRPA. Clinical laboratories should evaluate their internal processes to ensure they are able to accurately detect and subsequently report CRPA. Some recommendations for consideration are provided in Table 3. It is important to remember that continuous communication and collaboration between clinical laboratories, their facilities’ infection prevention partners, their local or state PHLs, and the CDC helps strengthen efforts for combatting antimicrobial resistance.REFERENCES
1. Centers for Disease Control. Outbreak of Extensively Drug-resistant Pseudomonas aeruginosa Associated with Artificial Tears. https://emergency.cdc.gov/han/2023/han00485.asp. Published February 1, 2023. Accessed April 27, 2023.
2. US Food and Drug Administration. FDA warns consumers not to purchase or use EzriCare Artificial Tears due to potential contamination. https://www.fda.gov/drugs/drug-safety-and-availability/fda-warns-consumers-not-purchase-or-use-ezricare-artificial-tears-due-potential-contamination. Published February 2, 2023. Accessed April 27, 2023.
3. Reyes J, Komarow L, Chen L, et al. Antibacterial Resistance Leadership Group and Multi-Drug Resistant Organism Network Investigators. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe. 2023 Mar;4(3):e159-e170.
4. Giani T, Arena F, Pollini S, et al. Pseudomonas aeruginosa Working Group. Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J Antimicrob Chemother. 2018 Mar 1;73(3):664-671.
5. Centers for Disease Control. Pseudomonas aeruginosa in Healthcare Settings. https://www.cdc.gov/hai/organisms/pseudomonas.html. Published November 13, 2019. Accessed April 27, 2023.
6. Centers for Disease Control Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed April 27, 2023.
7. World Health Organization Media Center. WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed Published February 27, 2019. Accessed April 20, 2023.
8. CDC Antimicrobial Resistance Laboratory Network. https://arpsp.cdc.gov/profile/arln/crpa. Accessed April 18, 2023.
9. Wang M, Earley M, Chen L, et al. Multi-Drug Resistant Organism Network Investigators. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis. 2022 Mar;22(3):401-412.
10. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 33rd ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2023.
11. Vallabhaneni S, Huang JY, Grass JE, et al. Antimicrobial Susceptibility Profiles to Predict the Presence of Carbapenemase Genes among Carbapenem-Resistant Pseudomonas aeruginosa Isolates. J Clin Microbiol. 2021 May 19;59(6):e02874-20.
12. Tamma PD, Simner PJ. Phenotypic Detection of Carbapenemase-Producing Organisms from Clinical Isolates. J Clin Microbiol. 2018 Oct 25;56(11):e01140-18.
13. Tenover FC. Using Molecular Diagnostics to Develop Therapeutic Strategies for Carbapenem-Resistant Gram-Negative Infections. Front Cell Infect Microbiol. 2021 Sep 28;11:715821.
14. Jenkins S, Ledeboer NA, Westblade LF, et al. Evaluation of NG-Test Carba 5 for Rapid Phenotypic Detection and Differentiation of Five Common Carbapenemase Families: Results of a Multicenter Clinical Evaluation. J Clin Microbiol. 2020 Jun 24;58(7):e00344-20.