For a printable version of the June CE Story and test go HERE or to take test online go HERE. For more information, visit the Continuing Education tab.
LEARNING OBJECTIVES
Upon completion of this article, the reader will be able to:
1. List and describe COVID-19 variants that have swept through the US population.
2. Discuss the study performed in LA County and Riverside County as it relates to: participants, samples transportation, validation and useability.
3. Describe results and finding of the study performed in LA County and Riverside County.
4. Discuss how future variants will be tracked and ways to keep the spread under control.
Like many viruses, coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), mutates.1 When mutations occur in key proteins, the virus can become more transmissible, become resistant to certain treatments, and gain the ability to evade antibody-mediated immunity. When that happens, the strain may be classified as a variant of concern.2,3
Following the initial reports of SARS-CoV-2 infections in China and Italy, the United States was not prepared for a pandemic of a novel respiratory virus. The initial SARS-CoV-2 detection assay developed by the US Centers for Disease Control and Protection had performance problems,4 the supply-chain for clinical diagnostic supplies was inadequate.5 and the public health system was neither prepared for the volume of SARS-CoV-2 testing nor surveillance for SARS-CoV-2.6 To have effectively responded to SARS-CoV-2, the rapid expansion of diagnostic testing and variant surveillance was needed early in the pandemic period.7
Following the initial wave of the wildtype strain of SARS-CoV-2, the first identified variant of concern was the Alpha variant (UK variant), which spread approximately 50% faster than the wildtype strain.2 Following the Alpha variant of concern, the Beta (South Africa), Gamma (Brazilian), Delta and Omicron variants of concern were identified.8,9 Currently, the Omicron variant with its sublineages is the most prevalent SARS-CoV-2 variant of concern in the United States,10,11 with new variants actively being identified.12
As evidenced by the Omicron variant,13 the transmissibility and ability to evade antibody-mediated immune protection can increase. Due to the major public health concern of SARS-CoV-2 and identified variants, we used a private SARS-CoV-2 clinical laboratory’s infrastructure and newly developed clinical research and sequencing capacity to monitor SARS-CoV-2 variant frequency and distribution in two large counties in California.
Participant characteristics
We enrolled adults (18 years or older) in Los Angeles County and Riverside County who recently tested positive for SARS-CoV-2 by PCR (Curative, San Dimas, CA). Participants were required to have a positive SARS-CoV-2 RT-PCR result with a cycle threshold value less than or equal to 30 cycles within 5 days of enrollment and sample collection. Trained healthcare workers instructed participants to self-collect anterior nares specimens under direct observation. Individuals meeting eligibility criteria and providing written consent were enrolled in the study.
Subjects considered vulnerable including pregnant women, nursing home residents or other institutionalized people, prisoners, and persons without decisional capacity were excluded from the study. Written informed consent was obtained from each eligible participant prior to enrollment in the study and any specimen collection. The study was approved by Advarra Institutional Review Board under Pro00053729 on May 10, 2021. All research was performed in accordance with relevant regulations in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants.
Sample transportation
Specimens were placed into 10 mL collection tubes containing DNA/RNA Shield Stabilization Solution (Zymo Research, Irvine, CA). Samples were transported to the sequencing laboratory at 2°C to 8°C within 5 hours of specimen collection and stored at 4°C for up to 7 days before library preparation.
Ribonucleic acid (RNA) isolation
RNA extraction was performed using chaotropic agents/silica-based methods. Either manual silica column-based extraction,33 or a modified automated magnetic silica beads-based extraction method were used. RNA was eluted in 60 mL of 10 mM Tris (pH 7.4). 34
Library preparation and sequencing
The libraries were prepared using COVIDSeq protocol.35 On a single 96-well plate, samples were processed alongside a positive control (SARS-CoV-2 BEI NR-52287 genotype A) and a negative control of human nasal specimen without SARS-CoV-2 RNA. Next, 96 indexed sample libraries from each plate were pooled together and quantified using a fluorometer. Four 96-well plates were combined at equimolar concentrations to a total of 384 samples and sequenced. Dilution and loading were performed as per the manufacturer’s instructions. Dual-indexed paired-end sequencing was performed for 100 cycles or 200 cycles to get a deeper sequencing depth. Sequencing aimed to have 1 to 2 million reads per sample.37
Quality control of reads
Paired-end reads were filtered and trimmed to reduce low quality base calls in the analysis and to eliminate the presence of primers and adapters. A minimum quality score of 30 was selected (pTrimer-1.3.4 -a Primers.bed -q 30 -t pair).14
Mapping
Paired-end reads were then mapped against the ‘Wuhan seafood market pneumonia virus isolate” Wuhan-Hu-1 genome (Accession number: NC_045512.2),15 using bwa.16 Each read was aligned (bwa aln -t 8 NC_045512.2.fasta), then alignments were paired with the sampe option. Alignment files were then subsetted using samtools view to consider only proper pairs with a quality score larger than 12 (samtools view -bS -q 12 -f 0X2).17
Validation
To validate the SARS-CoV-2 sequencing assay, BEI SARS-CoV-2 samples were used as control samples.18 Sequencing libraries were prepared by two operators. Each operator would prepare 16 replicates of BEI 52287, which would be used as the positive control in clinical sample testing, and one library for each of the other 13 BEI standards. All extracted nucleic acids from the BEI samples were verified to contain SARS-CoV-2 RNA. Ct values were between 26 and 29 for each sample before proceeding to cDNA synthesis. cDNA synthesis and library were prepared. Finished libraries were enzymatically normalized and pooled. Pooled libraries were quantified with KAPA qPCR and pooled together in equimolar concentrations to make a standardized final pooled library. 38 The final pooled library was sequenced and analyzed with the protocol listed later.
Variant calling and consensus genome generation, mutation identification, and variant identification
Variable sites were generated, regardless of coverage depth assuming a haploid genome using bcftools (bcftools call -mv -Ov).17 Variable sites were then filtered for quality (20) coverage (20x) and minimum allele frequency (0.25). Finally, the consensus genome was generated using the VCFcons.py script (VCFCons.py --input_depth TEMP.depth --input_vcf sample.vcf --vcf_type bcftools -c 10 -f 0.25 -q 20), part of a CoSa suite. 39 Consensus genomes were then run on the command line version of both: pangolin and nextclade.19
Ad-hoc analysis
Custom scripts were used to calculate sequencing, effort, and the percentage of reads used to assemble de novo genomes and base pair coverage. These can be found at (https://github.com/curative/bioinformatics).
Analysis of consensus sequences mutations and identifying similar isolates
A lineage comparison was done using Outbreak.info resource created by Scripps Research. A search for genomic sequences similar to the identified SARS-CoV-2 isolates was performed using Nucleotide BLAST 2.6.0+. Isolated were compared to all sequences available on the Global Influenza Surveillance & Response System (https://www.epicov.org/epi3/cfrontend#2c08bd). Data underwent alignment to identify gaps in generated consensus sequences and to match them with positions in amino acid sequences carrying hallmark mutations using software.40
Reporting
Results were submitted to Global Influenza Surveillance & Response System EpiCoV database for widespread data sharing and surveillance, a public surveillance service.20,21 The accession numbers were added to the Global Influenza Surveillance & Response System.
Useability
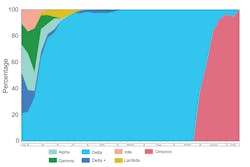
From May 27, 2021, to January 11, 2022, 820 recently tested SARS-CoV-2 positive participants were enrolled and underwent specimen collection. Of those enrolled, there were 408 (49.8%) females, 570 (69.5%) vaccinated, and 351 (42.8%) of Hispanic or Spanish origin. Median age of participant was 43 years (IQR: 33, 53). Of the cohort, 803 (97.9%) participants had symptoms at time of collection. The time from specimen collection to sequence result was reduced to three days. During the study period, we observed a decreased prevalence of Alpha, Gamma, Iota, Lambda, which was replaced by Delta, then Omicron variant of SARS-CoV-2 (Figure 1).
The view from here
In all, outpatient SARS-CoV-2 variant surveillance could be conducted by a private laboratory in a timely and accurate manner. Surveillance programs are needed to monitor SARS-CoV-2 variants to inform public health efforts. With the development of new genomic sequencing tools, it is possible for genomic data to be used to inform those public health responses. The World Health Organization proposed that a strong and resilient global sequencing network that provides useable and timely results is needed to maximize the public health impact of sequencing.22
The identification of new SARS-CoV-2 variants in a timely manner is critical to public health. While it is hard to prognosticate the future, it is possible to establish a method to prioritize research when new mutations are discovered on genetic coding segments of key proteins, like the SARS-CoV-2 spike protein.23,24 Faster identification of new SARS-CoV-2 variants of concern and understanding the rates in their change of prevalence could be critical predictors of new waves of SARS-CoV-2 and met with changes in public health recommendations. This study demonstrates that private clinical laboratories may play a role in the surveillance of SARS-CoV-2 variants of concern.
The sheer number of people who have been infected and the total SARS-CoV-2 infected person-time has led to the rapid evolution of SARS-CoV-2. Local epidemics of populous areas creates a situation in which many new mutations can form due to the large amount of viral spread over a short period of time. Additionally, there are many reports of SARS-CoV-2 detected among many animal species that closely interact with people, which may become reservoirs of infection and future spillover events.25
Animal reservoirs
As long as SARS-CoV-2 infections persist, SARS-CoV-2 will continue to mutate, and new variants of concern will arise. So far, farmed mink and pet hamsters have been shown to be capable of infecting humans with SARS-CoV-2.26 SARS-CoV-2 has also been identified among many domestic and wild animal species, e.g., bats, hamsters, ferrets, minks, cats, white-tailed deer, apes, and pigs.25 There is evidence that SARS-CoV-2 can be spread among various animal species and between animal species.27,28
Some research suggests that spillback of SARS-CoV-2 into other animal species has been observed with accelerating frequency with concerns of rapid adaption that may hasten viral evolution and novel strain emergence.29 Bashor, et al. observed rapid selection of SARS-CoV-2 variants in vitro and in vivo studies using cell-expanded SARS-CoV-2 inoculum and viruses recovered from cats, dogs, hamsters, and a ferret following experiment exposure. However, it is not clear how these animal reservoirs will contribute to endemic SARS-CoV-2 infections, mutation of new clinically significant variants of concern, or the risk of zoonotic spread. Given the severity of the pandemic caused by SARS-CoV-2, it seems prudent to not only monitor animal species known to harbor SARS-CoV-2 for presence of virus, but also for potentially dangerous mutations that could develop into new variants of concern.
Tracking future variants into the endemic phase
After the wave caused by Omicron variant of SARS-CoV-2 subsided, there has been a lower global prevalence of SARS-CoV-2 and many countries have rolled back public health measures used to prevent the spread of SARS-CoV-2.30 However, as Tedros Adhanom Ghebreyesus, the Director-General for the World Health Organization, noted, the world will be living with COVID-19 for the foreseeable future.30 While we now have the basic tools needed to address COVID-19—testing, treatment, and vaccinations, it is clear that areas of our surveillance, public health, and medical systems need to be bolstered. Vaccines importantly continue to prevent severe infections, however vaccine effectiveness against infection wanes with time, new variants of concern can partially evade the immune system, and large populations around the world have not had adequate access vaccinations for SARS-CoV-2.31 While effective treatments have been developed for SARS-CoV-2, treatments are costly, have supply-chain issues for availability and distribution, and a lack of awareness of treatments causes them to be underutilized.32 While many testing modalities for SARS-CoV-2 which have been developed including rapid testing and genomic testing, access to COVID-19 testing continues to be a problem, especially among the uninsured. Frequent and routine testing should be made available for the public to help guide public health measures to address local epidemics, and continued genomic sequencing is needed to assess SARS-CoV-2 mutations to monitor new variants of concern.
Conclusions
The tools needed to address SARS-CoV-2 have been developed; however, continued vigilance is needed as the frequency and distribution of SARS-CoV-2 transitions from a pandemic to endemic state. This study demonstrates that timely outpatient SARS-CoV-2 variant surveillance conducted by a private laboratory could be used to inform public health efforts to identify changes in SARS-CoV-2 strains in local communities. Government agencies should engage private clinical laboratories in the surveillance of diseases that threaten the public’s health to supplement national disease surveillance networks.
References
- Baric RS. Emergence of a Highly Fit SARS-CoV-2 Variant. N Engl J Med. Dec 31 2020;383(27):2684-2686. doi:10.1056/NEJMcibr2032888. https://www.nejm.org/doi/pdf/10.1056/NEJMcibr2032888. Accessed April 18, 2022.
- Lauring AS, Malani PN. Variants of SARS-CoV-2. JAMA. Aug 13 2021;doi:10.1001/jama.2021.14181. https://jamanetwork.com/journals/jama/fullarticle/2783248. Accessed April 18, 2022.
- Tao K, Tzou PL, Nouhin J, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet. Sep 17 2021;doi:10.1038/s41576-021-00408-x. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8447121/. Accessed April 18, 2022.
- Schneider EC. Failing the Test - The Tragic Data Gap Undermining the U.S. Pandemic Response. N Engl J Med. Jul 23 2020;383(4):299-302. doi:10.1056/NEJMp2014836. https://www.nejm.org/doi/full/10.1056/NEJMp2014836. Accessed April 18, 2022.
- Mirchandani P. Health Care Supply Chains: COVID-19 Challenges and Pressing Actions. Ann Intern Med. Aug 18 2020;173(4):300-301. doi:10.7326/M20-1326. https://pubmed.ncbi.nlm.nih.gov/32369540/. Accessed April 18, 2022.
- Maxmen A. Has COVID taught us anything about pandemic preparedness? Nature. Aug 2021;596(7872):332-335. doi:10.1038/d41586-021-02217-y. https://www.nature.com/articles/d41586-021-02217-y. Accessed April 18, 2022.
- Krijger PHL, Hoek TA, Boersma S, et al. A public-private partnership model for COVID-19 diagnostics. Nat Biotechnol. Oct 2021;39(10):1182-1184. doi:10.1038/s41587-021-01080-6. https://search.bvsalud.org/global-literature-on-novel-coronavirus-2019-ncov/resource/es/covidwho-1442791. Accessed April 18, 2022.
- Abdool Karim SS, de Oliveira T. New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N Engl J Med. May 13 2021;384(19):1866-1868. doi:10.1056/NEJMc2100362. https://www.nejm.org/doi/10.1056/NEJMc2100362. Accessed April 18, 2022.
- Del Rio C, Malani PN, Omer SB. Confronting the Delta Variant of SARS-CoV-2, Summer 2021. JAMA. Aug 18 2021;doi:10.1001/jama.2021.14811. https://jamanetwork.com/journals/jama/fullarticle/2783478. Accessed April 18, 2022.
- Jansen L, Tegomoh B, Lange K, et al. Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) Variant Cluster - Nebraska, November-December 2021. MMWR Morb Mortal Wkly Rep. Dec 31 2021;70(5152):1782-1784. doi:10.15585/mmwr.mm705152e3. https://www.cdc.gov/mmwr/volumes/70/wr/mm705152e3.htm. Accessed April 18, 2022.
- Team CC-R. SARS-CoV-2 B.1.1.529 (Omicron) Variant - United States, December 1-8, 2021. MMWR Morb Mortal Wkly Rep. Dec 17 2021;70(50):1731-1734. doi:10.15585/mmwr.mm7050e1. https://pubmed.ncbi.nlm.nih.gov/34914670/. Accessed April 18, 2022.
- Wink PL, Volpato FCZ, Monteiro FL, et al. First identification of SARS-CoV-2 Lambda (C.37) variant in Southern Brazil. Infect Control Hosp Epidemiol. Sep 2 2021:1-7. doi:10.1017/ice.2021.390. https://www.cambridge.org/core/services/aop-cambridge-core/content/view/42A50C4938E81BAAE1F4CAF1E03B9419/S0899823X21003901a.pdf/div-class-title-first-identification-of-sars-cov-2-lambda-c-37-variant-in-southern-brazil-div.pdf. Accessed April 18, 2022.
- Willyard C. What the Omicron wave is revealing about human immunity. Nature. Feb 2022;602(7895):22-25. doi:10.1038/d41586-022-00214-3. https://www.nature.com/articles/d41586-022-00214-3. Accessed April 18, 2022.
- Zhang X, Shao Y, Tian J, et al. pTrimmer: An efficient tool to trim primers of multiplex deep sequencing data. BMC Bioinformatics. May 10 2019;20(1):236. doi:10.1186/s12859-019-2854-x. https://www.semanticscholar.org/paper/pTrimmer%3A-An-efficient-tool-to-trim-primers-of-deep-Zhang-Shao/93234c66e21ee3ce0d543120e05faa5bfb7ec921. Accessed April 18, 2022.
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. Mar 2020;579(7798):265-269. doi:10.1038/s41586-020-2008-3. https://www.nature.com/articles/s41586-020-2008-3. Accessed April 18, 2022.
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. Mar 1 2010;26(5):589-95. doi:10.1093/bioinformatics/btp698. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2828108/. Accessed April 18, 2022.
- Danecek P, Bonfield JK, Liddle J, et al. Twelve years of SAMtools and BCFtools. Gigascience. Feb 16 2021;10(2)doi:10.1093/gigascience/giab008.
- Xun G, Lane ST, Petrov VA, Pepa BE, Zhao H. A rapid, accurate, scalable, and portable testing system for COVID-19 diagnosis. Nat Commun. May 18 2021;12(1):2905. doi:10.1038/s41467-021-23185-x. https://doaj.org/article/2694721fc8374a76a6fcc49618e6ec06. Accessed April 18, 2022.
- Rambaut A, Holmes EC, O’Toole A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. Nov 2020;5(11):1403-1407. doi:10.1038/s41564-020-0770-5. https://www.nature.com/articles/s41564-020-0770-5. Accessed April 18, 2022.
- Maxmen A. One million coronavirus sequences: popular genome site hits mega milestone. Nature. May 2021;593(7857):21. doi:10.1038/d41586-021-01069-w. https://www.nature.com/articles/d41586-021-01069-w. Accessed April 18, 2022.
- Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. Mar 30 2017;22(13)doi:10.2807/1560-7917.ES.2017.22.13.30494. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5388101/. Accessed April 18, 2022.
- Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health (World Health Organization) (2021). https://www.who.int/publications/i/item/9789240018440. Accessed April 18, 2022.
- Callaway E. The mutation that helps Delta spread like wildfire. Nature. Aug 2021;596(7873):472-473. doi:10.1038/d41586-021-02275-2. https://www.nature.com/articles/d41586-021-02275-2. Accessed April 18, 2022.
- Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. Jul 2021;19(7):409-424. doi:10.1038/s41579-021-00573-0. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8167834/. Accessed April 18, 2022.
- Mallapaty S. The search for animals harbouring coronavirus - and why it matters. Nature. Mar 2021;591(7848):26-28. doi:10.1038/d41586-021-00531-z. https://www.nature.com/articles/d41586-021-00531-z. Accessed April 18, 2022.
- Joint statement on the prioritization of monitoring SARS-CoV-2 infection in wildlife and preventing the formation of animal reservoirs (WHO) (2022). https://www.who.int/news/item/07-03-2022-joint-statement-on-the-prioritization-of-monitoring-sars-cov-2-infection-in-wildlife-and-preventing-the-formation-of-animal-reservoirs. Accessed April 18, 2022.
- Farag EA, Islam MM, Enan K, El-Hussein AM, Bansal D, Haroun M. SARS-CoV-2 at the human-animal interphase: A review. Heliyon. Dec 2021;7(12):e08496. doi:10.1016/j.heliyon.2021.e08496. https://europepmc.org/article/PMC/PMC8626158. Accessed April 18, 2022.
- Bonilla-Aldana DK, Rodriguez-Morales AJ. The threat of the spread of SARS-CoV-2 variants in animals. Vet Q. Dec 2021;41(1):321-322. doi:10.1080/01652176.2021.2008046. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8635604/. Accessed April 18, 2022.
- Bashor L, Gagne RB, Bosco-Lauth AM, Bowen RA, Stenglein M, VandeWoude S. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc Natl Acad Sci U S A. Nov 2 2021;118(44)doi:10.1073/pnas.2105253118. https://www.pnas.org/doi/10.1073/pnas.2105253118. Accessed April 18, 2022.
- Adepoju P. Africa prepares for endemic COVID-19. Nat Med. Mar 11 2022;doi:10.1038/d41591-022-00040-0. https://www.nature.com/articles/d41591-022-00040-0. Accessed April 18, 2022.
- Armstrong K. Covid-19 and the Investigator Pipeline. N Engl J Med. Jul 1 2021;385(1):7-9. doi:10.1056/NEJMp2100086. https://www.nejm.org/doi/full/10.1056/NEJMp2100086. Accessed April 18, 2022.
- Perwitasari DA, Faridah IN, Dania H, et al. The knowledge of COVID-19 treatments, behaviors, and attitudes of providing the information on COVID-19 treatments: Perspectives of pharmacy students. J Educ Health Promot. 2021;10:235. doi:10.4103/jehp.jehp_1416_20.
- Total RNA Purification Kit 96 Deep Well Plate Format Dx, Norgen Biotek Corp., Thorold, ON. https://norgenbiotek.com/. Accessed April 18, 2022.
- Thermo Scientific KingFisher Flex, Thermo Scientific, Waltham, MA. www.thermofisher.com/us/en/home.html. Accessed April 18, 2022.
- Illumina COVIDSeq protocol Illumina, Inc., San Diego, CA. www.illumina.com. Accessed April 18, 2022.
- Qubit 3.0, Invitrogen, Waltham, MA. www.thermofisher.com/us/en/home.html. Accessed April 18, 2022.
- NextSeq2000, Illumina, Inc., San Diego, CA. www.illumina.com. Accessed April 18, 2022.
- Roche KAPA qPCR, Indianapolis, IN. https://sequencing.roche.com/en-us.html. Accessed April 18, 2022.
- CoSa suite Pacific Biosciences, Menlo Park, CA. https://www.pacb.com/. Accessed April 18, 2022.
- Geneious Prime software, Geneious, Auckland, New Zealand. https://www.geneious.com/prime/. Accessed April 18, 2022.
Jeffrey D. Klausner, MD, MPH Clinical Professor of Medicine, Population and Public Health Sciences Keck School of Medicine of the University of Southern California.
Dr. Noah Kojima obtained his medical degree from the UCLA David Geffen School of Medicine, University of California Los Angeles, CA, where he also completed and lead the Global Health Track program.
Additional article contributors: Eugenia Khorosheva, Lauren Lopez, Mikhail Hanewich-Hollatz, J. Cesar Ignacio-Espinoza, Matthew Brobeck, Janet Chen, Matthew Geluz, Victoria Hess, Sophia Quasem, Nabjot Sandhu, Elias Salfati, Maria Shacreaw, George Way, Zhiyi Xie, Vladimir Slepnev