Group B Streptococcus is recognized as a leading cause of infectious early neonatal morbidity and mortality in the United States and around the world. Up to 30 percent of pregnant women are colonized with GBS during pregnancy, which can be transmitted to the infant at birth and may cause disease and death.1 Following the recommendations for screening pregnant woman for GBS colonization, the rate of early-onset GBS disease decreased from 1.8 cases per 1,000 live births to 0.23 cases per 1,000 live births.2
The recommendations to screen all pregnant women for GBS colonization was first released in 1996 by the American College of Obstetricians and Gynecologists (ACOG). This was followed by guidance from the Centers for Disease Control and Prevention (CDC) and the American Academy of Pediatrics (AAP). In 2019, the CDC granted stewardship of these guidelines to three professional organizations: ACOG and AAP are now responsible for guidelines on prophylaxis and treatment of GBS infection in pregnant women and newborns, and the American Society for Microbiology (ASM) is responsible for maintaining and updating guidelines for standard laboratory practices related to detection and identification of GBS during pregnancy.3
The guidelines from ASM recommend the following related to screening cultures for GBS during pregnancy:
- GBS screening specimens should be incubated in selective enrichment broth prior to agar media plating or nucleic acid amplification testing (NAAT).
- Culture media and GBS isolation methods should detect both hemolytic and nonhemolytic strains.
- Acceptable phenotypic and proteomic methods of identification of suspected isolates include CAMP test, latex agglutination and mass spectrometry.
- Nucleic acid amplification-based identification of GBS from enrichment broth is acceptable but not sufficient for all patients.
- Latex agglutination directly from enrichment broth and direct-from-specimen immunoassays are unacceptable methods for GBS detection.
According to ASM’s guidance, there is currently insufficient data to recommend direct-from-specimen testing by NAAT, emphasizing that culture after broth enrichment should remain a hallmark step in GBS screening.
Laboratory screening for GBS in pregnant women
Since the release of this initial guidance for GBS culture screening of pregnant women, researchers have been looking for more efficient and sensitive methods to detect GBS colonization. Several investigators have studied the sensitivity of NAAT testing after broth-enrichment, as compared to conventional culture showing acceptable results.4,5,6 The use of chromogenic agars has also provided some advances in the screening process. Salem and Anderson performed direct culture of vaginal specimens to four different chromogenic media and showed that these agars produced comparable results to conventional broth-enrichment sub-cultured to routine blood agar plates.7 Church et. al. compared their standard broth (carrot broth) sub-cultured to blood agar (with a neomycin disk) to chromogenic agar and to NAAT testing.8 Their results showed that NAAT and the chromogenic agar cultures had the same performance with sensitivities, specificities, and positive and negative predictive values of 98.4 percent, 99.6 percent, 98.4 percent, and 99.6 percent, respectively, as compared to their routine procedure. Likewise, El Aila et. al. showed that direct plating to chromogenic agar provided rapid detection of GBS that was as sensitive and specific as the recommended method of broth sub-cultured onto nonchromogenic agar.9
Laboratory automation and intelligent software algorithms in microbiology
Automation is becoming increasingly popular in microbiology laboratories with systems that include smart incubators (providing constant atmospheric and temperature conditions for cultures) plus digital imaging systems to record images of plates at user-specified times. These digital images are then viewed by technologists for culture work-up rather than handling the plates themselves. Recently, a suite of advanced artificial intelligence (AI) algorithms with the ability to pre-assess and pre-sort culture plates based on its ‘reading’ of digital plate images has become available, and previous studies have shown the ability of this software to increase the sensitivity and specificity for the detection of Methicillin-resistant Staphylococcus aureus (MRSA), Vancomycin-resistant Enterococci (VRE) and Group A Streptococcus using chromogenic media.10,11,12 For example, the study by Faron, et.al, tested over 100,000 specimens for the presence of VRE and showed that the software agreed with manual digital plate reading for 90.1 percent of all specimens resulting in a software sensitivity and specificity of 100 percent and 89.5 percent, respectively.10 The software never called a positive result incorrectly as negative, but, in fact, it detected an additional 499 positive specimens that were falsely called negative by the technologist. Overall, the software was 100 percent accurate when it identified VRE-negative plates.
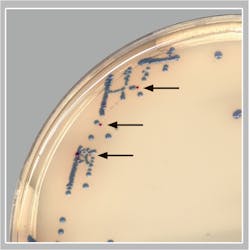
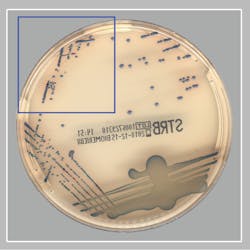
The remainder of this article will focus on a recent publication by Baker et. al. showing the ability of AI algorithms to detect GBS from pregnancy screening cultures when using a specifically designed chromogenic agar.13 They evaluated the clinical performance of a digital imaging software algorithm in the detection of GBS from broth-enriched specimens plated on a new chromogenic media. The chromogenic agar used in the study was a selective and differential medium, which shows GBS organisms as pink to red colonies that are round and pearly on clear agar. The software was used to evaluate images of colonies for the appropriate coloration on the chromogenic agar, indicating a possible GBS organism. The software then would pre-sort culture plates based on its ‘reading’ of digital plate images. For example, the software recognized and differentiated colony colors on chromogenic agars and sorted the cultures into “negative for GBS” and “potential positive for GBS.” Over 650 vaginal-rectal swabs enriched in broth cultures were enrolled at two sites, automatically processed, and sent to smart incubators with digital imaging after 24 and 48 hours of incubation. The performance of the software (SW) for detecting GBS on the chromogenic agar was compared to technologist manual review (MR) of the chromogenic agar digital images and to NAAT.
The prominent findings from this research were:
- MR had a sensitivity and specificity of 84.5 percent and 94.7 percent after 48 hours of incubation as compared to NAAT.
- SW had a sensitivity of 100 percent, as compared to MR with no true positive isolates missed by 48 hours of incubation.
- Of the specimens that were negative by MR and positive by the SW, an additional 8 true positive GBS specimens were detected (see Figure).
- When comparing all of the methods for performance and adjudication of discordant results, the sensitivities of the MR, SW, and NAAT were 90.3 percent, 95.5 percent, and 96.8 percent, respectively. The specificities of the MR, SW, and NAAT were 95.4 percent, 63.0 percent, and 97.7 percent, respectively.
Cost comparison between NAAT and digital cultures with software reading
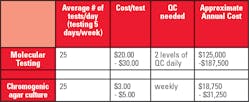
The authors discuss that for those laboratories that continue to culture for GBS, a total of 36-48 hours is required to obtain results, along with a significant amount of manual labor. For those that have turned to NAAT to detect GBS, the gains in sensitivity and workflow are used to justify the increased cost of the molecular methods. It should be noted that routine GBS screening is recommended to be performed within five weeks of birth, and therefore, when performing routine screening, the rapid turnaround time offered by NAAT is not required. The table shows the authors’ estimated annual cost for NAAT versus chromogenic agar culture. A GBS NAAT assay would likely range from $20-$30 per test with 2 levels of QC needed daily. In a laboratory performing 25 tests per day for 5 days/week (approximately 250 days/year), the reagent cost of the molecular testing would be $125,000-$187,500 /year. Conversely, with the cost of chromogenic agar at approximately $3-5 per plate and needed quality control performed only weekly, the same laboratory would spend between $18,750 - $31,250 annually for cultures. The authors conclude that with the increasing implementation of automation in the laboratory, intelligent software algorithms that do not sacrifice sensitivity, increase the opportunity to use the less expensive culture-based methods that recently have been replaced by NAAT.Summary
There is no doubt that the use of artificial intelligence in clinical microbiology has only just begun. Increased automation with algorithm-aided culture ‘reading’ can handle mundane tasks, leaving the technologist free to perform more important duties. This and other recent publications show us that automation with digital imaging and intelligent software algorithms can revitalize culture in our clinical microbiology laboratories.
References:
- Shabayek S, Spellerberg B. Group B Streptococcal Colonization, Molecular Characteristics, and Epidemiology. Front Microbiol. 2018; 9:437. DOI: 10.3389/fmicb.2018.00437.
- Prevention of Group B Streptococcal early-onset disease in newborns. Committee Opinion. American College of Obstetrics and Gynecologists. https://www.acog.org/clinical/clinical-guidance/committee-opinion/articles/2020/02/prevention-of-group-b-streptococcal-early-onset-disease-in-newborns. Accessed December 1, 2020.
- Filkins L, Hauser J, Robinson-Dunn B et al. Guidelines for the detection and identification of Group B Streptococcus. American Society for Microbiology. https://asm.org/ASM/media/Policy-and-Advocacy/images/ASM-GBS-guideline-031020.pdf?ext=.pdf. Accessed December 1, 2020.
- Shin JH, Pride DT. Comparison of three nucleic acid amplification tests and culture for detection of group B Streptococcus from enrichment broth. J Clin Microbiol. 2019; 57(6):1-9. doi: 10.1128/JCM.01958-18.
- Hernandez DR, Wolk DM, Walker KL, et. al. Multicenter diagnostic accuracy evaluation of the luminex aries real-time PCR assay for group B streptococcus detection in LIM broth-enriched samples. J Clin Microbiol. 218; 56(8):1-9. doi: 10.1128/JCM.01768-17.
- Buchan BW, Faron ML, Fuller D, Davis TE, Mayne D, Ledeboer NA. Multicenter clinical evaluation of the xpert GBS LB assay for detection of group b streptococcus in prenatal screening specimens. J Clin Microbiol. 2015; 53(2):443–448. doi: 10.1128/JCM.02598-14.
- Salem N, Anderson JJ. Evaluation of four chromogenic media for the isolation of Group B Streptococcus from vaginal specimens in pregnancy women. Pathology. 2015; 47(6): 580-582. doi: 10.1097/PAT.0000000000000299.
- Church DL, Baxter H, Lloyd T, Larios O, Gregson DB. Evaluation of StrepBSelect chromogenic medium and the Fast-Track Diagnostics group B streptococcus (GBS) realtime PCR assay compared to routine culture for detection of GBS during antepartum screening. J Clin Microbiol. 2017; 55(7):2137–2142. doi: 10.1128/JCM.00043-17.
- El Aila NA, Tency I, Claeys G, et al. Comparison of different sampling techniques and of different culture methods for detection of group B streptococcus carriage in pregnant women. BMC Infect Dis. 2010;10:285. doi:10.1186/1471-2334-10-285.
- Faron ML, Buchan BW, Coon C, et. al. Automatic digital analysis of chromogenic media for Vancomycin-Resistant-Enterococcus screens using Copan WASPLab. J Clin Microbiol. 2016; 54(10): 2464-2469. doi: 10.1128/JCM.01040-16.
- Faron ML, Buchan BW, Vismara C, et al. Automated scoring of chromogenic media for detection of Methicillin-Resistant Staphylococcus aureus by use of WASPLab image analysis software. J Clin Microbiol. 2016; 54(3): 620-624. doi: 10.1128/JCM.02778-15.
- Van TT, Mata K, Dien Bard J. Automated detection of Streptococcus pyogenes pharyngitis using Colorex Strep A CHROMagar and WASPLab artificial intelligence chromogenic detection module software. J Clin Microbiol. 2019; 57(11): e00811-19. DOI: 10.1128/JCM.00811-19.
- Baker J, Timm K, Faron M, Ledeboer N, Culbreath K. Digital image analysis for the detection of Group B Streptococcus from ChromID StreptoB media using a phenomatrix artificial intelligence software algorithm. J Clin Microbiol. 2020 Oct 21:JCM.01902-19. doi: 10.1128/JCM.01902-19. Epub ahead of print.